Sulaiman Vesal
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020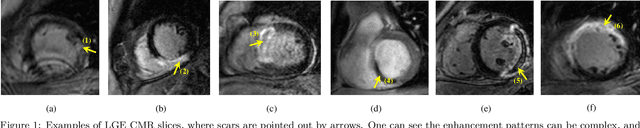
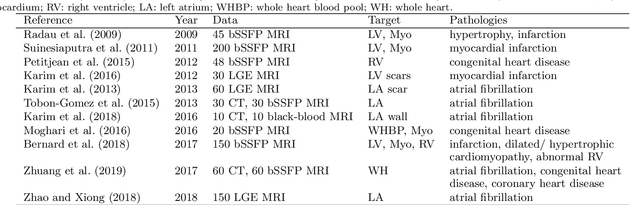
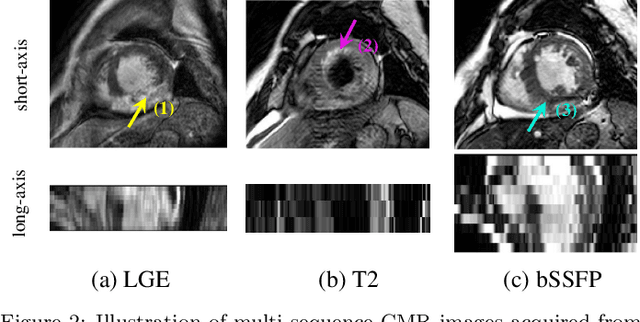
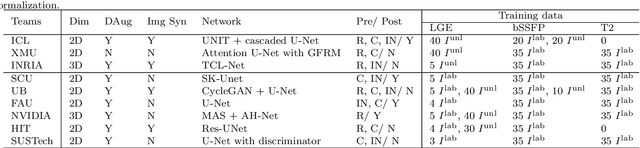
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020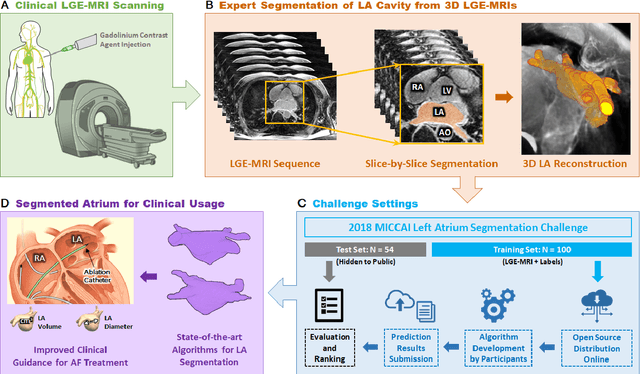
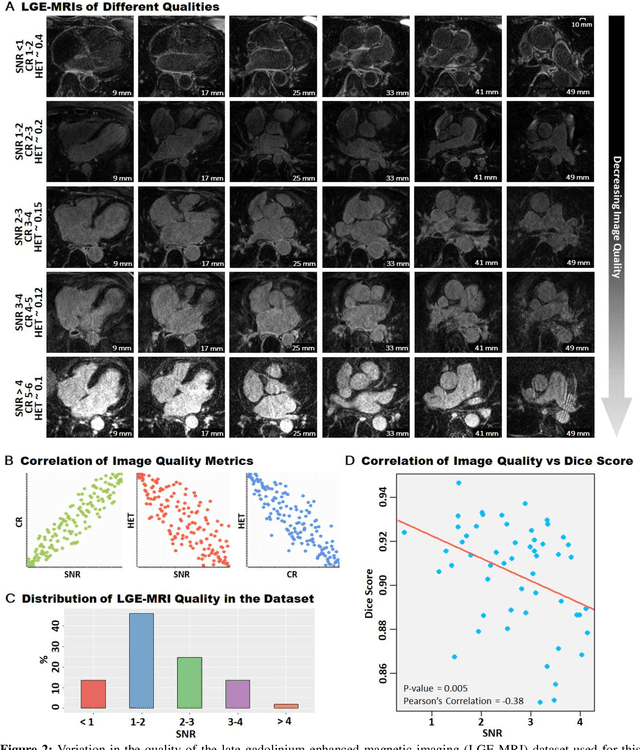
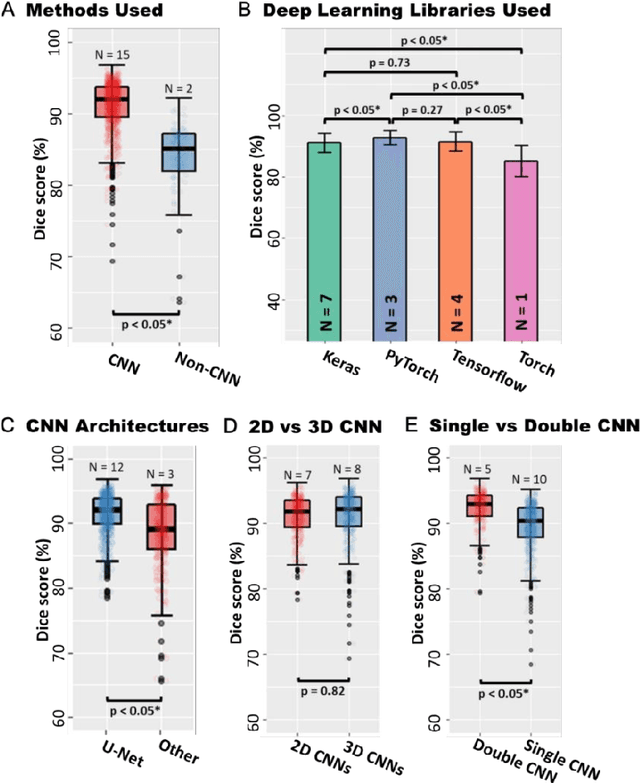
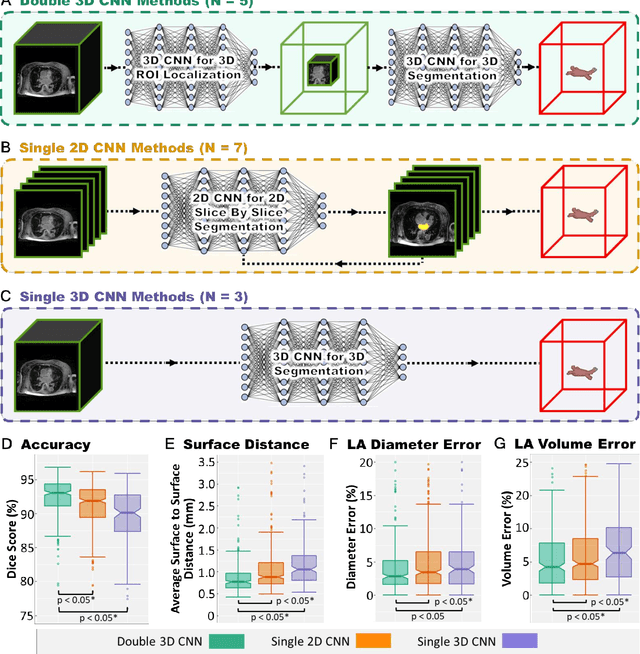
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
The Effect of Data Augmentation on Classification of Atrial Fibrillation in Short Single-Lead ECG Signals Using Deep Neural Networks
Feb 13, 2020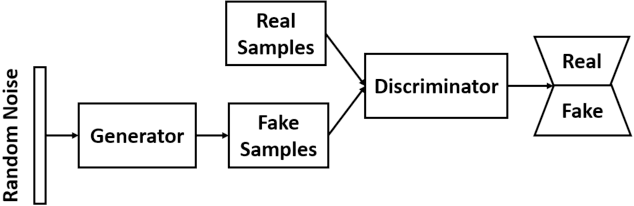
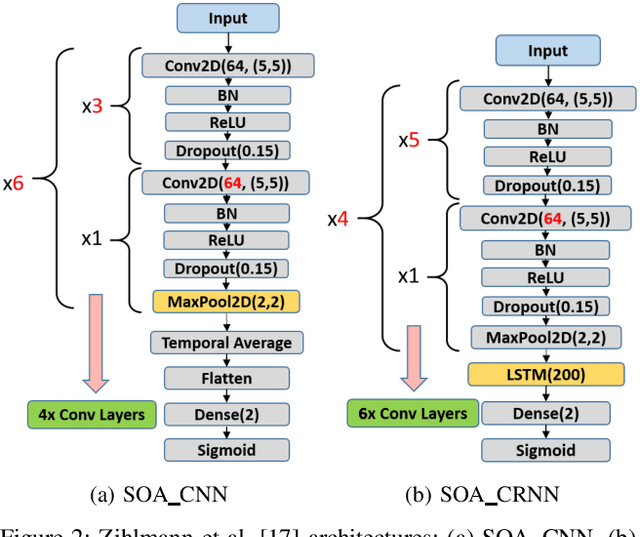
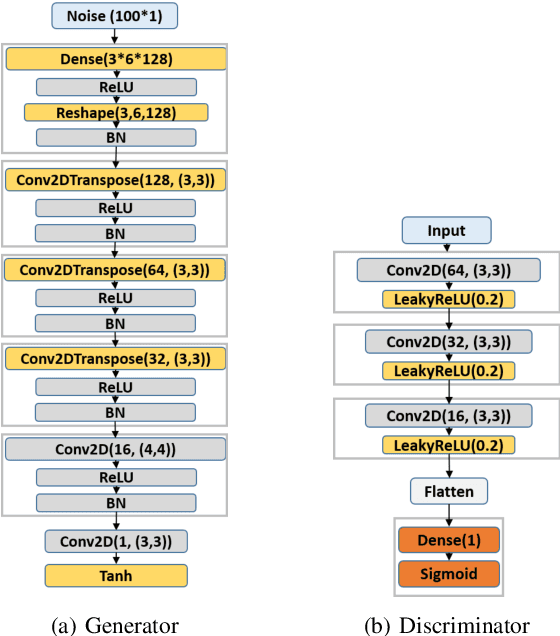
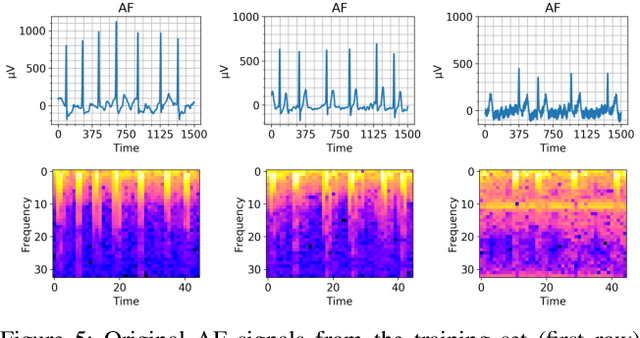
Abstract:Cardiovascular diseases are the most common cause of mortality worldwide. Detection of atrial fibrillation (AF) in the asymptomatic stage can help prevent strokes. It also improves clinical decision making through the delivery of suitable treatment such as, anticoagulant therapy, in a timely manner. The clinical significance of such early detection of AF in electrocardiogram (ECG) signals has inspired numerous studies in recent years, of which many aim to solve this task by leveraging machine learning algorithms. ECG datasets containing AF samples, however, usually suffer from severe class imbalance, which if unaccounted for, affects the performance of classification algorithms. Data augmentation is a popular solution to tackle this problem. In this study, we investigate the impact of various data augmentation algorithms, e.g., oversampling, Gaussian Mixture Models (GMMs) and Generative Adversarial Networks (GANs), on solving the class imbalance problem. These algorithms are quantitatively and qualitatively evaluated, compared and discussed in detail. The results show that deep learning-based AF signal classification methods benefit more from data augmentation using GANs and GMMs, than oversampling. Furthermore, the GAN results in circa $3\%$ better AF classification accuracy in average while performing comparably to the GMM in terms of f1-score.
COPD Classification in CT Images Using a 3D Convolutional Neural Network
Jan 04, 2020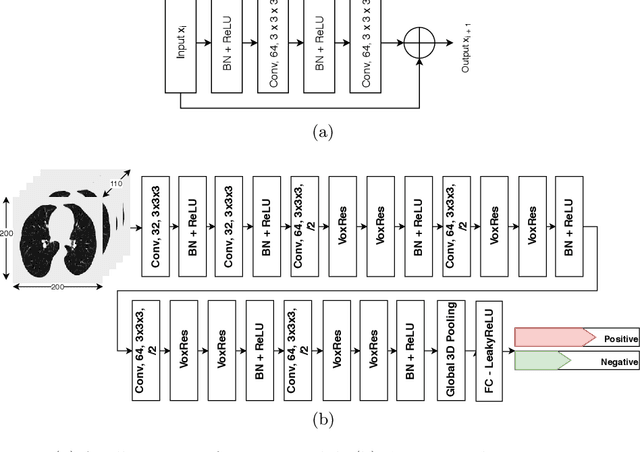
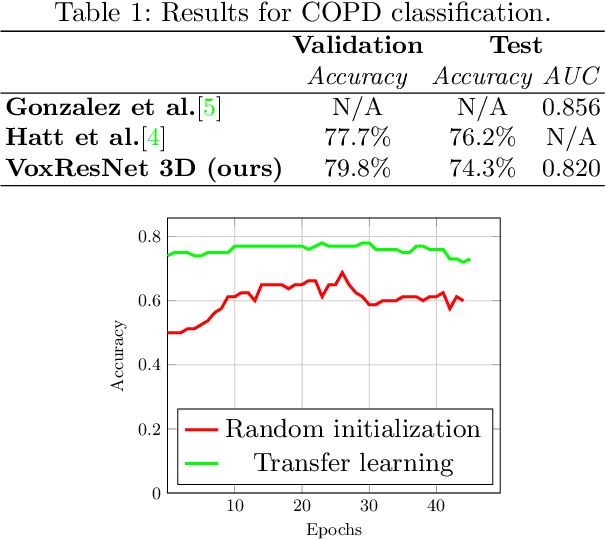
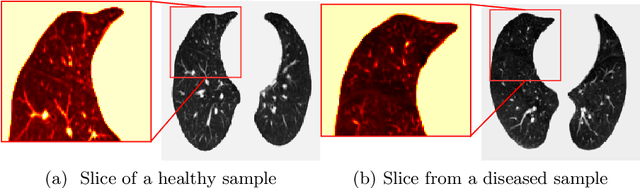
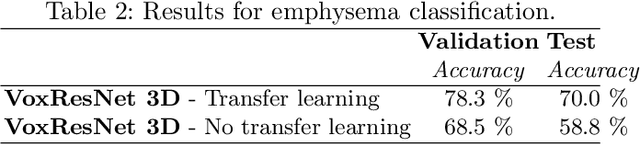
Abstract:Chronic obstructive pulmonary disease (COPD) is a lung disease that is not fully reversible and one of the leading causes of morbidity and mortality in the world. Early detection and diagnosis of COPD can increase the survival rate and reduce the risk of COPD progression in patients. Currently, the primary examination tool to diagnose COPD is spirometry. However, computed tomography (CT) is used for detecting symptoms and sub-type classification of COPD. Using different imaging modalities is a difficult and tedious task even for physicians and is subjective to inter-and intra-observer variations. Hence, developing meth-ods that can automatically classify COPD versus healthy patients is of great interest. In this paper, we propose a 3D deep learning approach to classify COPD and emphysema using volume-wise annotations only. We also demonstrate the impact of transfer learning on the classification of emphysema using knowledge transfer from a pre-trained COPD classification model.
Deep Learning-based Denoising of Mammographic Images using Physics-driven Data Augmentation
Dec 11, 2019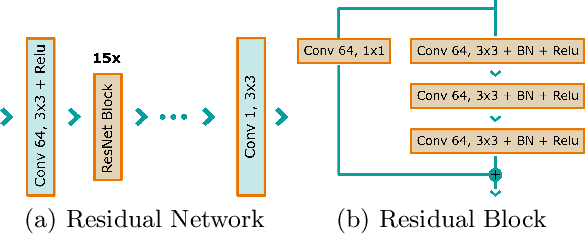

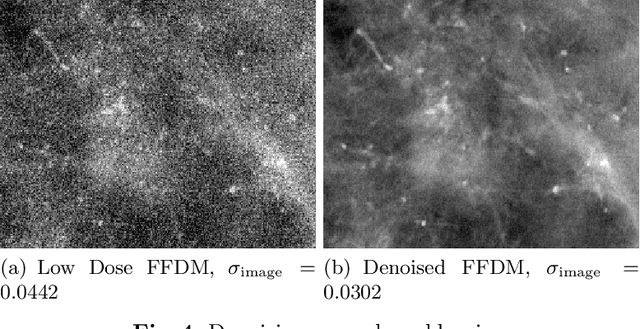
Abstract:Mammography is using low-energy X-rays to screen the human breast and is utilized by radiologists to detect breast cancer. Typically radiologists require a mammogram with impeccable image quality for an accurate diagnosis. In this study, we propose a deep learning method based on Convolutional Neural Networks (CNNs) for mammogram denoising to improve the image quality. We first enhance the noise level and employ Anscombe Transformation (AT) to transform Poisson noise to white Gaussian noise. With this data augmentation, a deep residual network is trained to learn the noise map of the noisy images. We show, that the proposed method can remove not only simulated but also real noise. Furthermore, we also compare our results with state-of-the-art denoising methods, such as BM3D and DNCNN. In an early investigation, we achieved qualitatively better mammogram denoising results.
Automated Multi-sequence Cardiac MRI Segmentation Using Supervised Domain Adaptation
Aug 21, 2019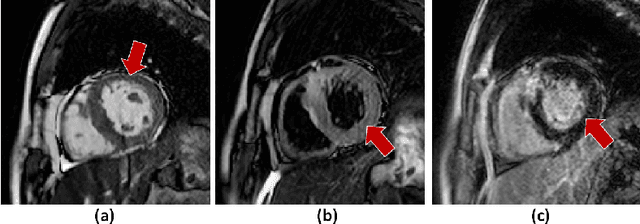
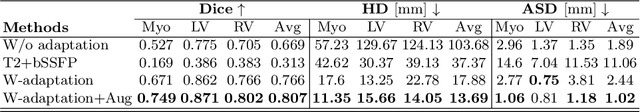
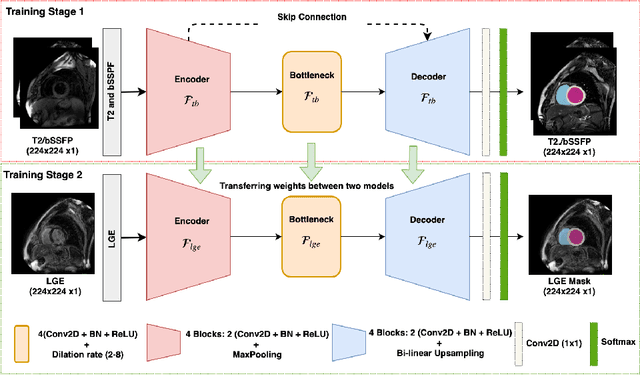
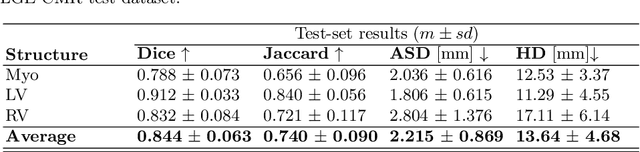
Abstract:Left ventricle segmentation and morphological assessment are essential for improving diagnosis and our understanding of cardiomyopathy, which in turn is imperative for reducing risk of myocardial infarctions in patients. Convolutional neural network (CNN) based methods for cardiac magnetic resonance (CMR) image segmentation rely on supervision with pixel-level annotations, and may not generalize well to images from a different domain. These methods are typically sensitive to variations in imaging protocols and data acquisition. Since annotating multi-sequence CMR images is tedious and subject to inter- and intra-observer variations, developing methods that can automatically adapt from one domain to the target domain is of great interest. In this paper, we propose an approach for domain adaptation in multi-sequence CMR segmentation task using transfer learning that combines multi-source image information. We first train an encoder-decoder CNN on T2-weighted and balanced-Steady State Free Precession (bSSFP) MR images with pixel-level annotation and fine-tune the same network with a limited number of Late Gadolinium Enhanced-MR (LGE-MR) subjects, to adapt the domain features. The domain-adapted network was trained with just four LGE-MR training samples and obtained an average Dice score of $\sim$85.0\% on the test set comprises of 40 LGE-MR subjects. The proposed method significantly outperformed a network without adaptation trained from scratch on the same set of LGE-MR training data.
A 2D dilated residual U-Net for multi-organ segmentation in thoracic CT
May 19, 2019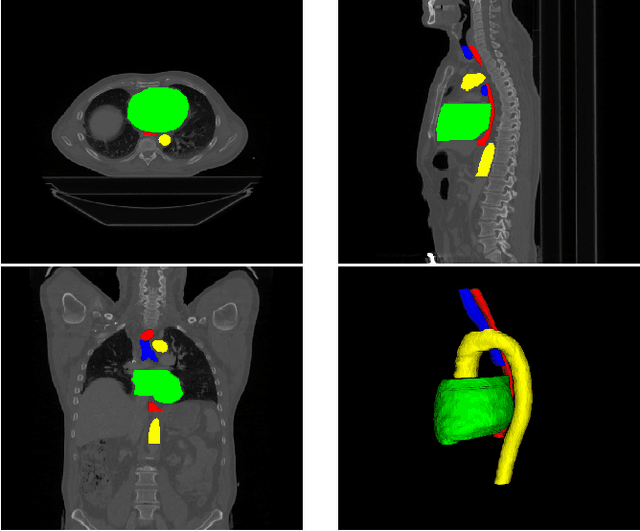
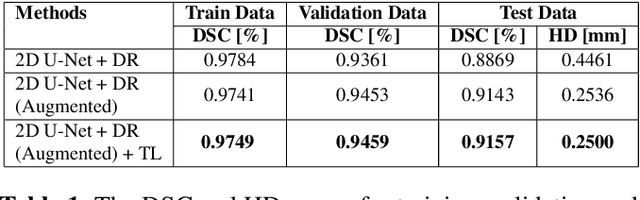
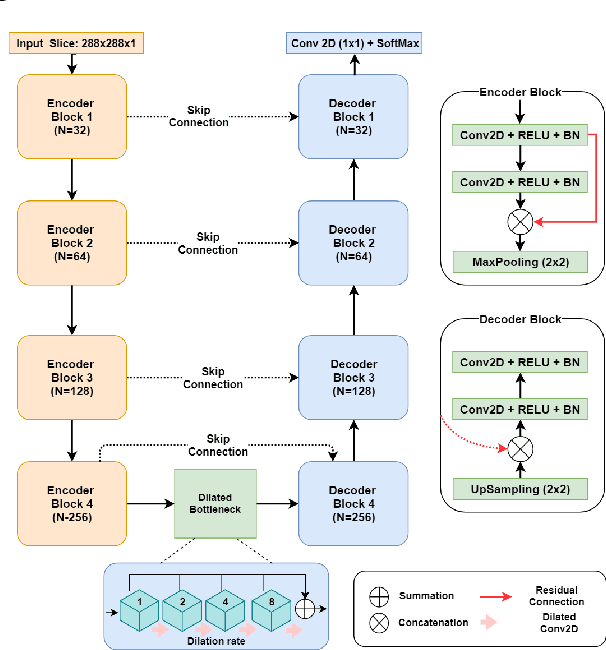

Abstract:Automatic segmentation of organs-at-risk (OAR) in computed tomography (CT) is an essential part of planning effective treatment strategies to combat lung and esophageal cancer. Accurate segmentation of organs surrounding tumours helps account for the variation in position and morphology inherent across patients, thereby facilitating adaptive and computer-assisted radiotherapy. Although manual delineation of OARs is still highly prevalent, it is prone to errors due to complex variations in the shape and position of organs across patients, and low soft tissue contrast between neighbouring organs in CT images. Recently, deep convolutional neural networks (CNNs) have gained tremendous traction and achieved state-of-the-art results in medical image segmentation. In this paper, we propose a deep learning framework to segment OARs in thoracic CT images, specifically for the: heart, esophagus, trachea and aorta. Our approach employs dilated convolutions and aggregated residual connections in the bottleneck of a U-Net styled network, which incorporates global context and dense information. Our method achieved an overall Dice score of 91.57% on 20 unseen test samples from the ISBI 2019 SegTHOR challenge.
Dilated deeply supervised networks for hippocampus segmentation in MRI
Mar 20, 2019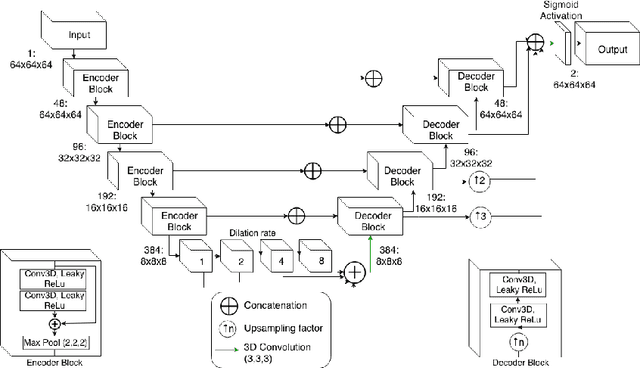
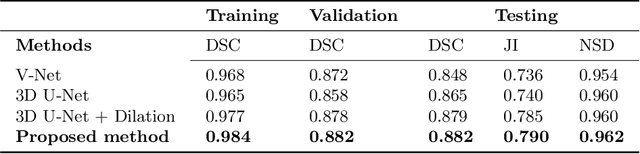
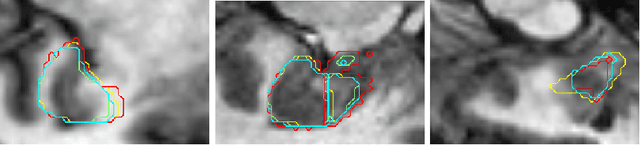
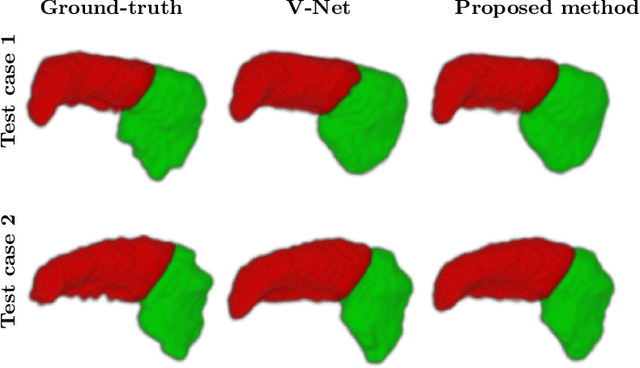
Abstract:Tissue loss in the hippocampi has been heavily correlated with the progression of Alzheimer's Disease (AD). The shape and structure of the hippocampus are important factors in terms of early AD diagnosis and prognosis by clinicians. However, manual segmentation of such subcortical structures in MR studies is a challenging and subjective task. In this paper, we investigate variants of the well known 3D U-Net, a type of convolution neural network (CNN) for semantic segmentation tasks. We propose an alternative form of the 3D U-Net, which uses dilated convolutions and deep supervision to incorporate multi-scale information into the model. The proposed method is evaluated on the task of hippocampus head and body segmentation in an MRI dataset, provided as part of the MICCAI 2018 segmentation decathlon challenge. The experimental results show that our approach outperforms other conventional methods in terms of different segmentation accuracy metrics.
A Multi-task Framework for Skin Lesion Detection and Segmentation
Aug 05, 2018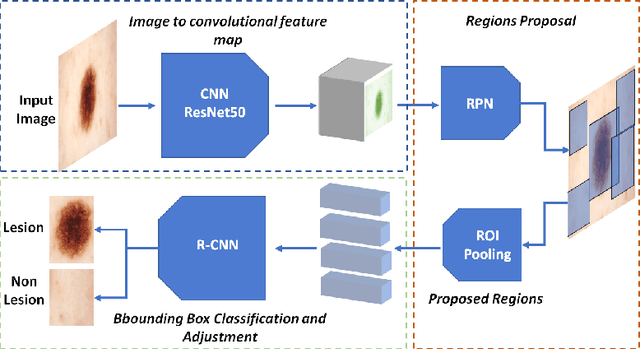

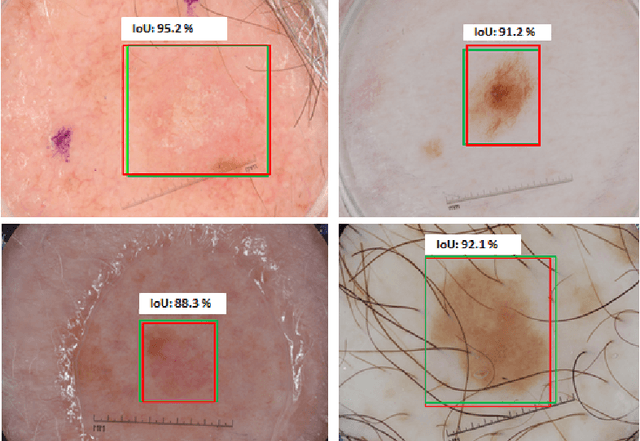
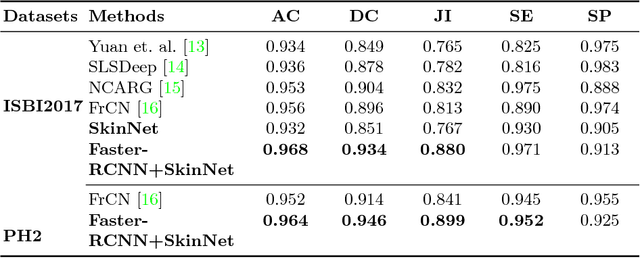
Abstract:Early detection and segmentation of skin lesions is crucial for timely diagnosis and treatment, necessary to improve the survival rate of patients. However, manual delineation is time consuming and subject to intra- and inter-observer variations among dermatologists. This underlines the need for an accurate and automatic approach to skin lesion segmentation. To tackle this issue, we propose a multi-task convolutional neural network (CNN) based, joint detection and segmentation framework, designed to initially localize the lesion and subsequently, segment it. A `Faster region-based convolutional neural network' (Faster-RCNN) which comprises a region proposal network (RPN), is used to generate bounding boxes/region proposals, for lesion localization in each image. The proposed regions are subsequently refined using a softmax classifier and a bounding-box regressor. The refined bounding boxes are finally cropped and segmented using `SkinNet', a modified version of U-Net. We trained and evaluated the performance of our network, using the ISBI 2017 challenge and the PH2 datasets, and compared it with the state-of-the-art, using the official test data released as part of the challenge for the former. Our approach outperformed others in terms of Dice coefficients ($>0.93$), Jaccard index ($>0.88$), accuracy ($>0.96$) and sensitivity ($>0.95$), across five-fold cross validation experiments.
Dilated Convolutions in Neural Networks for Left Atrial Segmentation in 3D Gadolinium Enhanced-MRI
Aug 05, 2018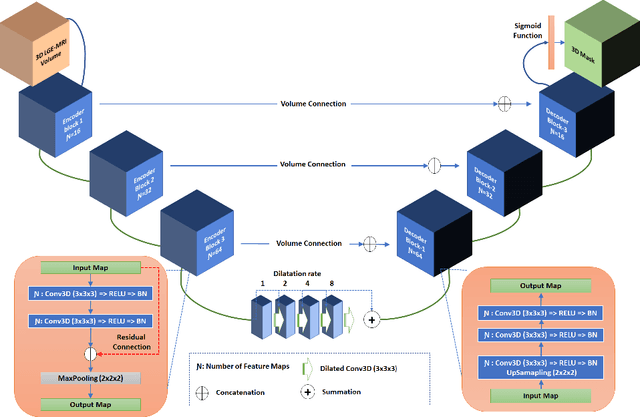
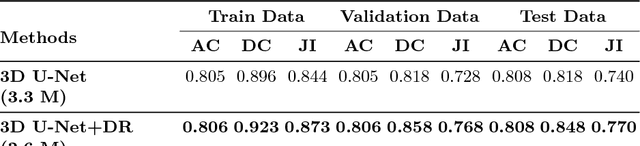
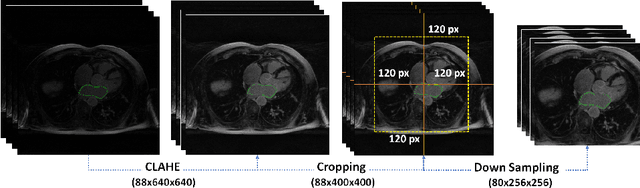
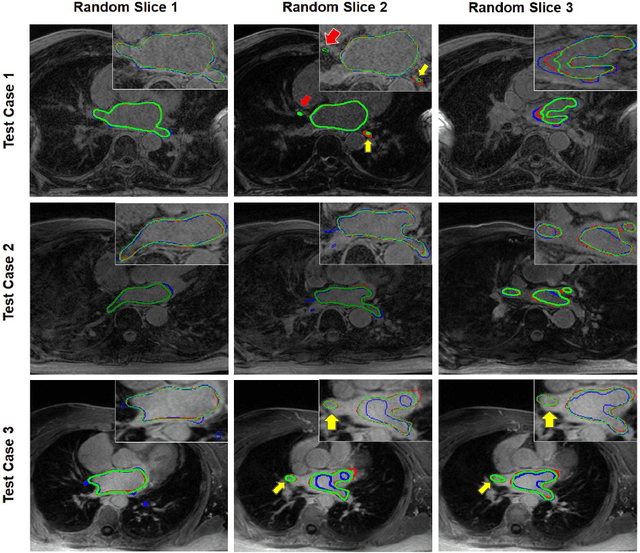
Abstract:Segmentation of the left atrial chamber and assessing its morphology, are essential for improving our understanding of atrial fibrillation, the most common type of cardiac arrhythmia. Automation of this process in 3D gadolinium enhanced-MRI (GE-MRI) data is desirable, as manual delineation is time-consuming, challenging and observer-dependent. Recently, deep convolutional neural networks (CNNs) have gained tremendous traction and achieved state-of-the-art results in medical image segmentation. However, it is difficult to incorporate local and global information without using contracting (pooling) layers, which in turn reduces segmentation accuracy for smaller structures. In this paper, we propose a 3D CNN for volumetric segmentation of the left atrial chamber in LGE-MRI. Our network is based on the well known U-Net architecture. We employ a 3D fully convolutional network, with dilated convolutions in the lowest level of the network, and residual connections between encoder blocks to incorporate local and global knowledge. The results show that including global context through the use of dilated convolutions, helps in domain adaptation, and the overall segmentation accuracy is improved in comparison to a 3D U-Net.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge