Oladimeji Farri
CVE-LLM : Ontology-Assisted Automatic Vulnerability Evaluation Using Large Language Models
Feb 21, 2025



Abstract:The National Vulnerability Database (NVD) publishes over a thousand new vulnerabilities monthly, with a projected 25 percent increase in 2024, highlighting the crucial need for rapid vulnerability identification to mitigate cybersecurity attacks and save costs and resources. In this work, we propose using large language models (LLMs) to learn vulnerability evaluation from historical assessments of medical device vulnerabilities in a single manufacturer's portfolio. We highlight the effectiveness and challenges of using LLMs for automatic vulnerability evaluation and introduce a method to enrich historical data with cybersecurity ontologies, enabling the system to understand new vulnerabilities without retraining the LLM. Our LLM system integrates with the in-house application - Cybersecurity Management System (CSMS) - to help Siemens Healthineers (SHS) product cybersecurity experts efficiently assess the vulnerabilities in our products. Also, we present guidelines for efficient integration of LLMs into the cybersecurity tool.
CVE-LLM : Automatic vulnerability evaluation in medical device industry using large language models
Jul 19, 2024



Abstract:The healthcare industry is currently experiencing an unprecedented wave of cybersecurity attacks, impacting millions of individuals. With the discovery of thousands of vulnerabilities each month, there is a pressing need to drive the automation of vulnerability assessment processes for medical devices, facilitating rapid mitigation efforts. Generative AI systems have revolutionized various industries, offering unparalleled opportunities for automation and increased efficiency. This paper presents a solution leveraging Large Language Models (LLMs) to learn from historical evaluations of vulnerabilities for the automatic assessment of vulnerabilities in the medical devices industry. This approach is applied within the portfolio of a single manufacturer, taking into account device characteristics, including existing security posture and controls. The primary contributions of this paper are threefold. Firstly, it provides a detailed examination of the best practices for training a vulnerability Language Model (LM) in an industrial context. Secondly, it presents a comprehensive comparison and insightful analysis of the effectiveness of Language Models in vulnerability assessment. Finally, it proposes a new human-in-the-loop framework to expedite vulnerability evaluation processes.
Fusion of Domain-Adapted Vision and Language Models for Medical Visual Question Answering
Apr 24, 2024Abstract:Vision-language models, while effective in general domains and showing strong performance in diverse multi-modal applications like visual question-answering (VQA), struggle to maintain the same level of effectiveness in more specialized domains, e.g., medical. We propose a medical vision-language model that integrates large vision and language models adapted for the medical domain. This model goes through three stages of parameter-efficient training using three separate biomedical and radiology multi-modal visual and text datasets. The proposed model achieves state-of-the-art performance on the SLAKE 1.0 medical VQA (MedVQA) dataset with an overall accuracy of 87.5% and demonstrates strong performance on another MedVQA dataset, VQA-RAD, achieving an overall accuracy of 73.2%.
General-Purpose vs. Domain-Adapted Large Language Models for Extraction of Data from Thoracic Radiology Reports
Dec 01, 2023



Abstract:Radiologists produce unstructured data that could be valuable for clinical care when consumed by information systems. However, variability in style limits usage. Study compares performance of system using domain-adapted language model (RadLing) and general-purpose large language model (GPT-4) in extracting common data elements (CDE) from thoracic radiology reports. Three radiologists annotated a retrospective dataset of 1300 thoracic reports (900 training, 400 test) and mapped to 21 pre-selected relevant CDEs. RadLing was used to generate embeddings for sentences and identify CDEs using cosine-similarity, which were mapped to values using light-weight mapper. GPT-4 system used OpenAI's general-purpose embeddings to identify relevant CDEs and used GPT-4 to map to values. The output CDE:value pairs were compared to the reference standard; an identical match was considered true positive. Precision (positive predictive value) was 96% (2700/2824) for RadLing and 99% (2034/2047) for GPT-4. Recall (sensitivity) was 94% (2700/2876) for RadLing and 70% (2034/2887) for GPT-4; the difference was statistically significant (P<.001). RadLing's domain-adapted embeddings were more sensitive in CDE identification (95% vs 71%) and its light-weight mapper had comparable precision in value assignment (95.4% vs 95.0%). RadLing system exhibited higher performance than GPT-4 system in extracting CDEs from radiology reports. RadLing system's domain-adapted embeddings outperform general-purpose embeddings from OpenAI in CDE identification and its light-weight value mapper achieves comparable precision to large GPT-4. RadLing system offers operational advantages including local deployment and reduced runtime costs. Domain-adapted RadLing system surpasses GPT-4 system in extracting common data elements from radiology reports, while providing benefits of local deployment and lower costs.
Generation of Radiology Findings in Chest X-Ray by Leveraging Collaborative Knowledge
Jun 18, 2023



Abstract:Among all the sub-sections in a typical radiology report, the Clinical Indications, Findings, and Impression often reflect important details about the health status of a patient. The information included in Impression is also often covered in Findings. While Findings and Impression can be deduced by inspecting the image, Clinical Indications often require additional context. The cognitive task of interpreting medical images remains the most critical and often time-consuming step in the radiology workflow. Instead of generating an end-to-end radiology report, in this paper, we focus on generating the Findings from automated interpretation of medical images, specifically chest X-rays (CXRs). Thus, this work focuses on reducing the workload of radiologists who spend most of their time either writing or narrating the Findings. Unlike past research, which addresses radiology report generation as a single-step image captioning task, we have further taken into consideration the complexity of interpreting CXR images and propose a two-step approach: (a) detecting the regions with abnormalities in the image, and (b) generating relevant text for regions with abnormalities by employing a generative large language model (LLM). This two-step approach introduces a layer of interpretability and aligns the framework with the systematic reasoning that radiologists use when reviewing a CXR.
* Information Technology and Quantitative Management (ITQM 2023)
shs-nlp at RadSum23: Domain-Adaptive Pre-training of Instruction-tuned LLMs for Radiology Report Impression Generation
Jun 05, 2023



Abstract:Instruction-tuned generative Large language models (LLMs) like ChatGPT and Bloomz possess excellent generalization abilities, but they face limitations in understanding radiology reports, particularly in the task of generating the IMPRESSIONS section from the FINDINGS section. They tend to generate either verbose or incomplete IMPRESSIONS, mainly due to insufficient exposure to medical text data during training. We present a system which leverages large-scale medical text data for domain-adaptive pre-training of instruction-tuned LLMs to enhance its medical knowledge and performance on specific medical tasks. We show that this system performs better in a zero-shot setting than a number of pretrain-and-finetune adaptation methods on the IMPRESSIONS generation task, and ranks 1st among participating systems in Task 1B: Radiology Report Summarization at the BioNLP 2023 workshop.
* 1st Place in Task 1B: Radiology Report Summarization at BioNLP 2023
RadLing: Towards Efficient Radiology Report Understanding
Jun 04, 2023



Abstract:Most natural language tasks in the radiology domain use language models pre-trained on biomedical corpus. There are few pretrained language models trained specifically for radiology, and fewer still that have been trained in a low data setting and gone on to produce comparable results in fine-tuning tasks. We present RadLing, a continuously pretrained language model using Electra-small (Clark et al., 2020) architecture, trained using over 500K radiology reports, that can compete with state-of-the-art results for fine tuning tasks in radiology domain. Our main contribution in this paper is knowledge-aware masking which is a taxonomic knowledge-assisted pretraining task that dynamically masks tokens to inject knowledge during pretraining. In addition, we also introduce an knowledge base-aided vocabulary extension to adapt the general tokenization vocabulary to radiology domain.
* Association for Computational Linguistics (ACL), 2023
Differentiable Multi-Agent Actor-Critic for Multi-Step Radiology Report Summarization
Mar 15, 2022
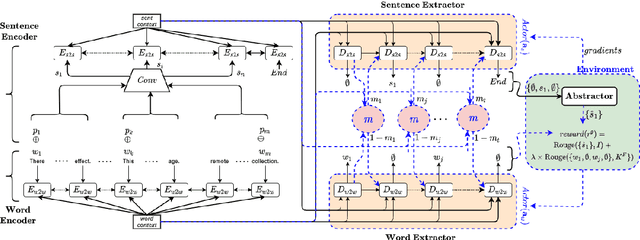

Abstract:The IMPRESSIONS section of a radiology report about an imaging study is a summary of the radiologist's reasoning and conclusions, and it also aids the referring physician in confirming or excluding certain diagnoses. A cascade of tasks are required to automatically generate an abstractive summary of the typical information-rich radiology report. These tasks include acquisition of salient content from the report and generation of a concise, easily consumable IMPRESSIONS section. Prior research on radiology report summarization has focused on single-step end-to-end models -- which subsume the task of salient content acquisition. To fully explore the cascade structure and explainability of radiology report summarization, we introduce two innovations. First, we design a two-step approach: extractive summarization followed by abstractive summarization. Second, we additionally break down the extractive part into two independent tasks: extraction of salient (1) sentences and (2) keywords. Experiments on a publicly available radiology report dataset show our novel approach leads to a more precise summary compared to single-step and to two-step-with-single-extractive-process baselines with an overall improvement in F1 score Of 3-4%.
* Accepted at 60th Annual Meeting of the Association for Computational Linguistics 2022 Main Conference
Assertion Detection in Multi-Label Clinical Text using Scope Localization
May 19, 2020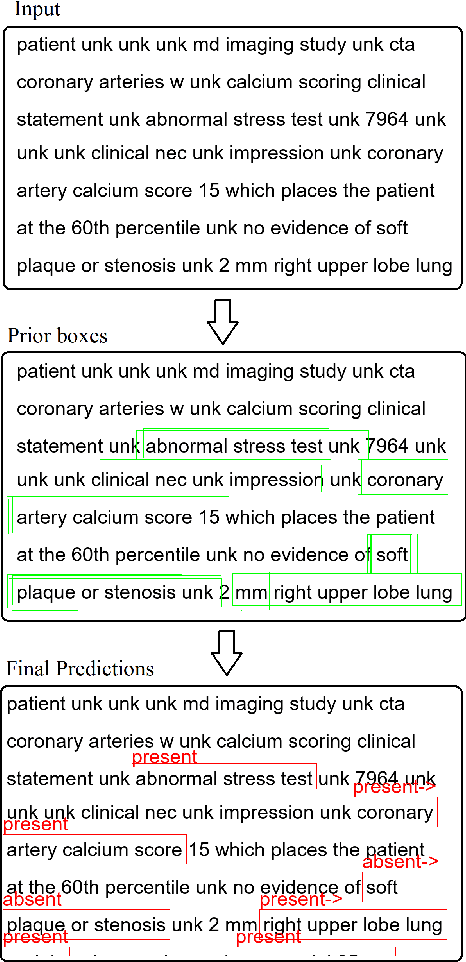
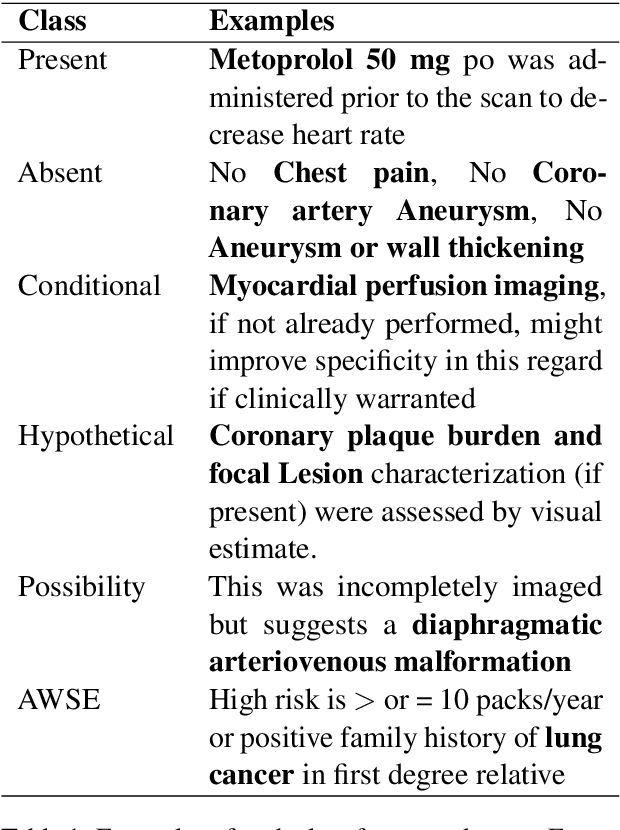
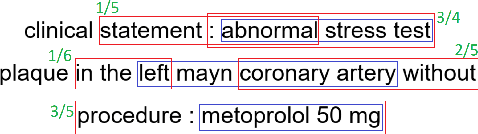
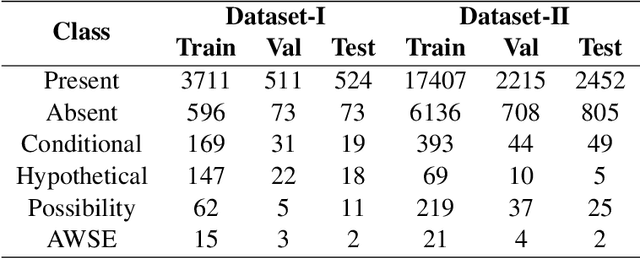
Abstract:Multi-label sentences (text) in the clinical domain result from the rich description of scenarios during patient care. The state-of-theart methods for assertion detection mostly address this task in the setting of a single assertion label per sentence (text). In addition, few rules based and deep learning methods perform negation/assertion scope detection on single-label text. It is a significant challenge extending these methods to address multi-label sentences without diminishing performance. Therefore, we developed a convolutional neural network (CNN) architecture to localize multiple labels and their scopes in a single stage end-to-end fashion, and demonstrate that our model performs atleast 12% better than the state-of-the-art on multi-label clinical text.
DR-BiLSTM: Dependent Reading Bidirectional LSTM for Natural Language Inference
Apr 11, 2018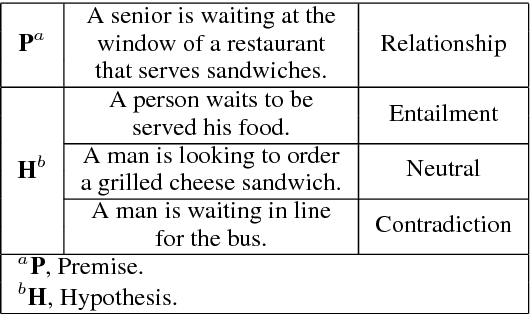
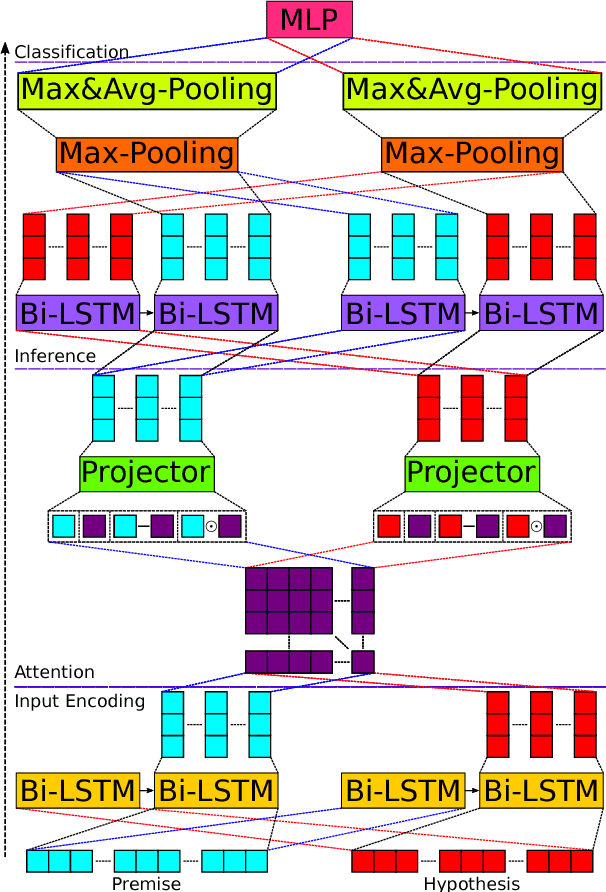
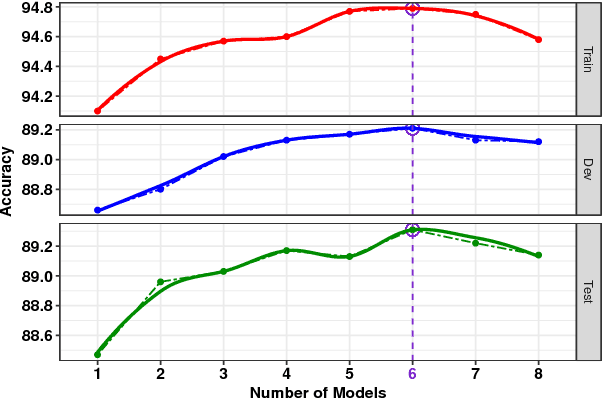
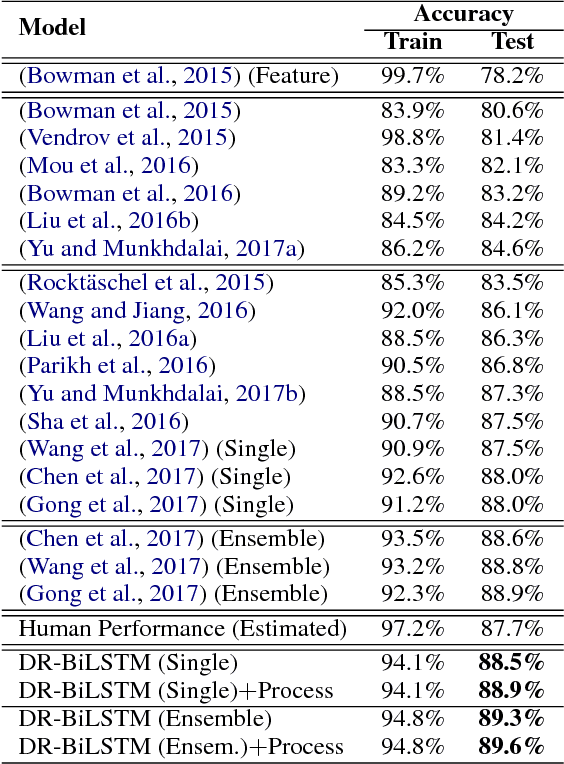
Abstract:We present a novel deep learning architecture to address the natural language inference (NLI) task. Existing approaches mostly rely on simple reading mechanisms for independent encoding of the premise and hypothesis. Instead, we propose a novel dependent reading bidirectional LSTM network (DR-BiLSTM) to efficiently model the relationship between a premise and a hypothesis during encoding and inference. We also introduce a sophisticated ensemble strategy to combine our proposed models, which noticeably improves final predictions. Finally, we demonstrate how the results can be improved further with an additional preprocessing step. Our evaluation shows that DR-BiLSTM obtains the best single model and ensemble model results achieving the new state-of-the-art scores on the Stanford NLI dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge