Sandy Engelhardt
Unsupervised Domain Adaptation from Axial to Short-Axis Multi-Slice Cardiac MR Images by Incorporating Pretrained Task Networks
Jan 20, 2021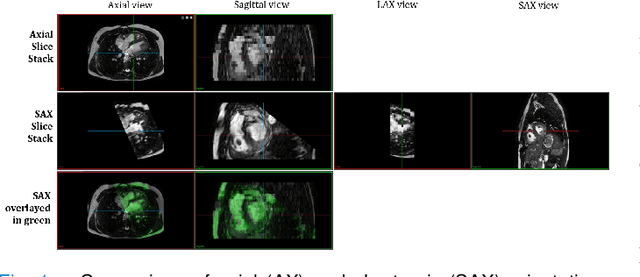
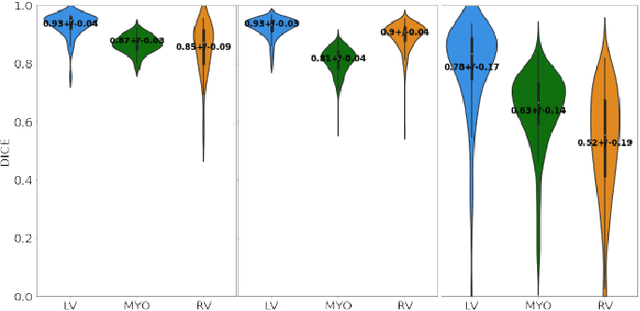
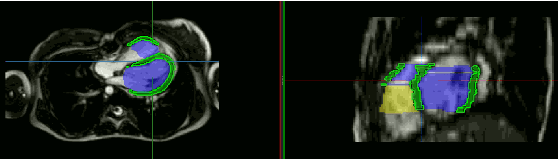
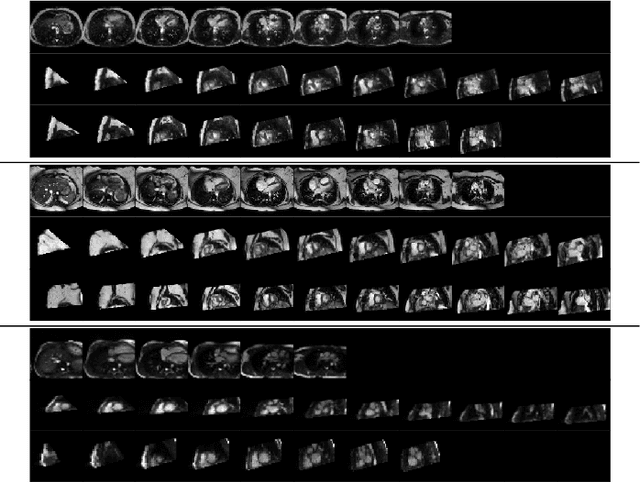
Abstract:Anisotropic multi-slice Cardiac Magnetic Resonance (CMR) Images are conventionally acquired in patient-specific short-axis (SAX) orientation. In specific cardiovascular diseases that affect right ventricular (RV) morphology, acquisitions in standard axial (AX) orientation are preferred by some investigators, due to potential superiority in RV volume measurement for treatment planning. Unfortunately, due to the rare occurrence of these diseases, data in this domain is scarce. Recent research in deep learning-based methods mainly focused on SAX CMR images and they had proven to be very successful. In this work, we show that there is a considerable domain shift between AX and SAX images, and therefore, direct application of existing models yield sub-optimal results on AX samples. We propose a novel unsupervised domain adaptation approach, which uses task-related probabilities in an attention mechanism. Beyond that, cycle consistency is imposed on the learned patient-individual 3D rigid transformation to improve stability when automatically re-sampling the AX images to SAX orientations. The network was trained on 122 registered 3D AX-SAX CMR volume pairs from a multi-centric patient cohort. A mean 3D Dice of $0.86\pm{0.06}$ for the left ventricle, $0.65\pm{0.08}$ for the myocardium, and $0.77\pm{0.10}$ for the right ventricle could be achieved. This is an improvement of $25\%$ in Dice for RV in comparison to direct application on axial slices. To conclude, our pre-trained task module has neither seen CMR images nor labels from the target domain, but is able to segment them after the domain gap is reduced. Code: https://github.com/Cardio-AI/3d-mri-domain-adaptation
Heatmap-based 2D Landmark Detection with a Varying Number of Landmarks
Jan 07, 2021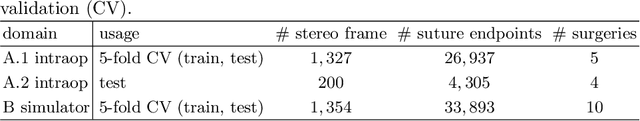
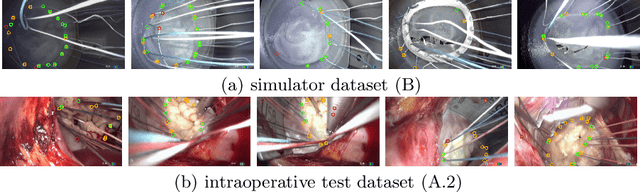
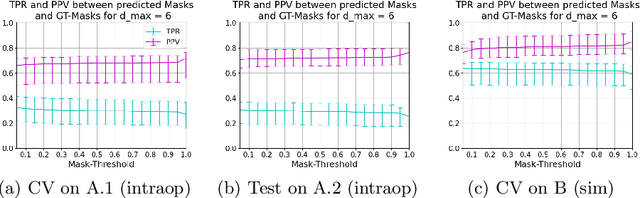
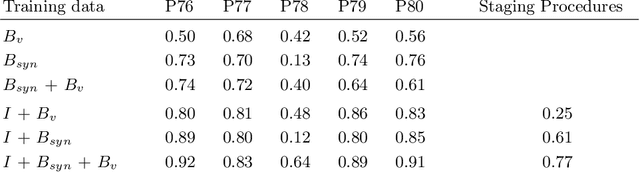
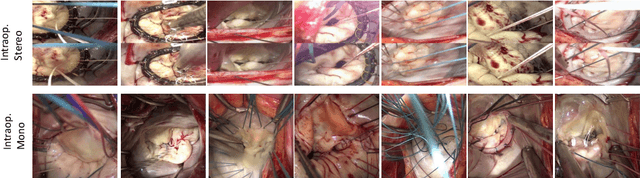
Abstract:Mitral valve repair is a surgery to restore the function of the mitral valve. To achieve this, a prosthetic ring is sewed onto the mitral annulus. Analyzing the sutures, which are punctured through the annulus for ring implantation, can be useful in surgical skill assessment, for quantitative surgery and for positioning a virtual prosthetic ring model in the scene via augmented reality. This work presents a neural network approach which detects the sutures in endoscopic images of mitral valve repair and therefore solves a landmark detection problem with varying amount of landmarks, as opposed to most other existing deep learning-based landmark detection approaches. The neural network is trained separately on two data collections from different domains with the same architecture and hyperparameter settings. The datasets consist of more than 1,300 stereo frame pairs each, with a total over 60,000 annotated landmarks. The proposed heatmap-based neural network achieves a mean positive predictive value (PPV) of 66.68$\pm$4.67% and a mean true positive rate (TPR) of 24.45$\pm$5.06% on the intraoperative test dataset and a mean PPV of 81.50\pm5.77\% and a mean TPR of 61.60$\pm$6.11% on a dataset recorded during surgical simulation. The best detection results are achieved when the camera is positioned above the mitral valve with good illumination. A detection from a sideward view is also possible if the mitral valve is well perceptible.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020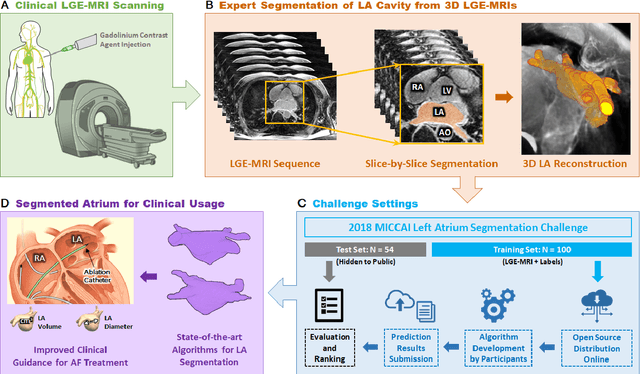
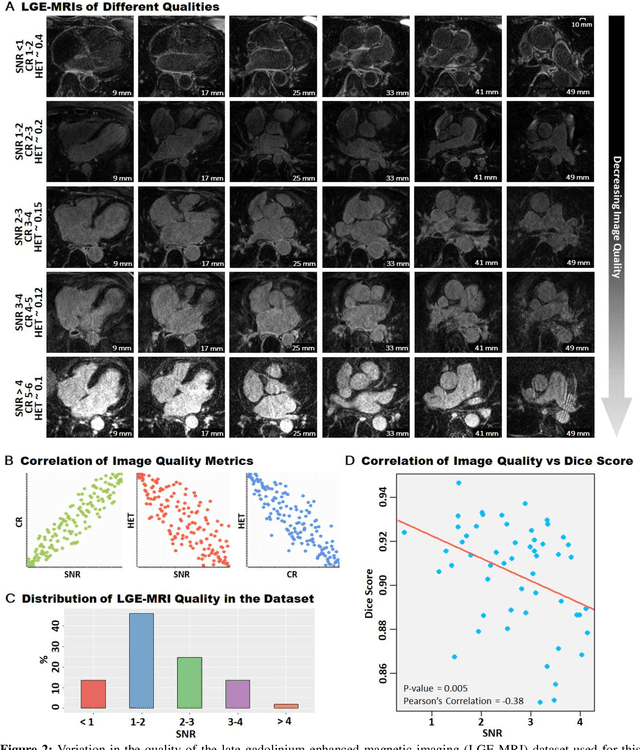
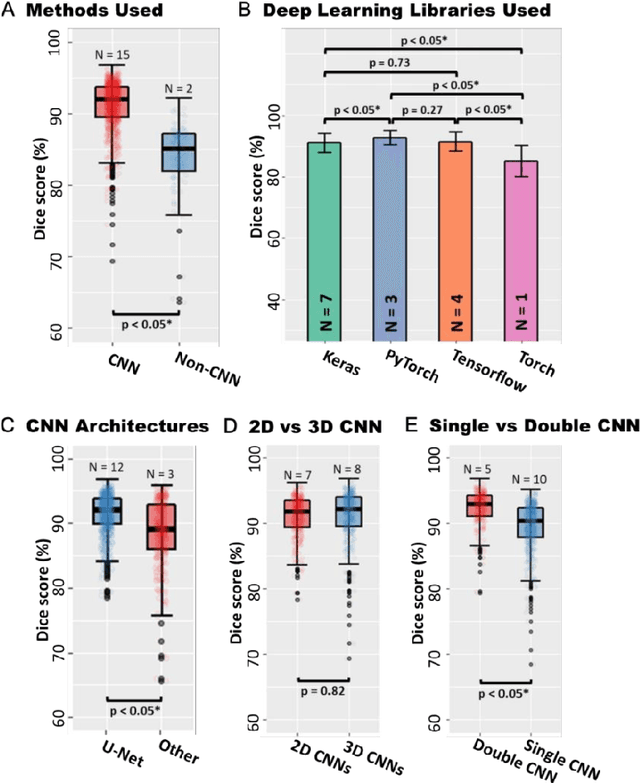
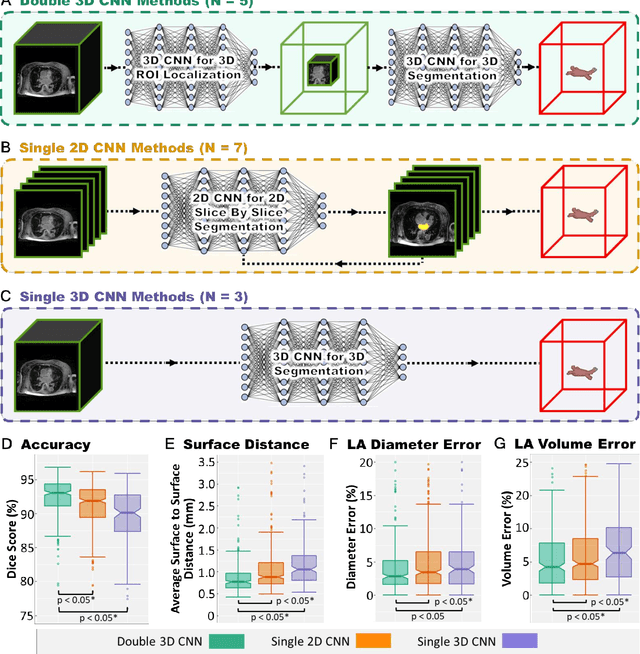
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
How well do U-Net-based segmentation trained on adult cardiac magnetic resonance imaging data generalise to rare congenital heart diseases for surgical planning?
Feb 10, 2020
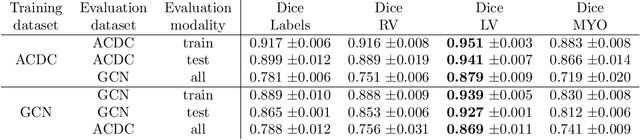
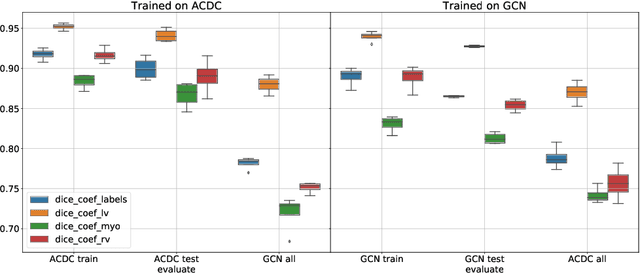
Abstract:Planning the optimal time of intervention for pulmonary valve replacement surgery in patients with the congenital heart disease Tetralogy of Fallot (TOF) is mainly based on ventricular volume and function according to current guidelines. Both of these two biomarkers are most reliably assessed by segmentation of 3D cardiac magnetic resonance (CMR) images. In several grand challenges in the last years, U-Net architectures have shown impressive results on the provided data. However, in clinical practice, data sets are more diverse considering individual pathologies and image properties derived from different scanner properties. Additionally, specific training data for complex rare diseases like TOF is scarce. For this work, 1) we assessed the accuracy gap when using a publicly available labelled data set (the Automatic Cardiac Diagnosis Challenge (ACDC) data set) for training and subsequent applying it to CMR data of TOF patients and vice versa and 2) whether we can achieve similar results when applying the model to a more heterogeneous data base. Multiple deep learning models were trained with four-fold cross validation. Afterwards they were evaluated on the respective unseen CMR images from the other collection. Our results confirm that current deep learning models can achieve excellent results (left ventricle dice of $0.951\pm{0.003}$/$0.941\pm{0.007}$ train/validation) within a single data collection. But once they are applied to other pathologies, it becomes apparent how much they overfit to the training pathologies (dice score drops between $0.072\pm{0.001}$ for the left and $0.165\pm{0.001}$ for the right ventricle).
Towards Augmented Reality-based Suturing in Monocular Laparoscopic Training
Jan 19, 2020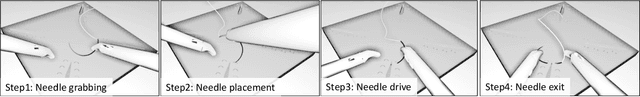
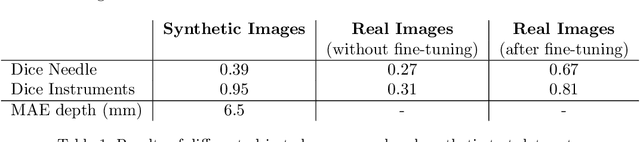
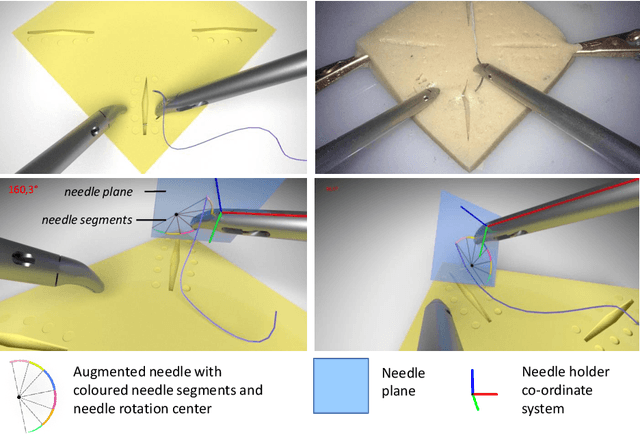
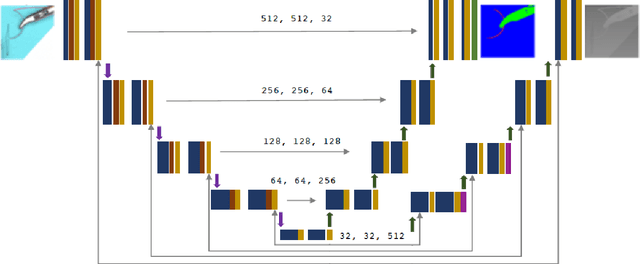
Abstract:Minimally Invasive Surgery (MIS) techniques have gained rapid popularity among surgeons since they offer significant clinical benefits including reduced recovery time and diminished post-operative adverse effects. However, conventional endoscopic systems output monocular video which compromises depth perception, spatial orientation and field of view. Suturing is one of the most complex tasks performed under these circumstances. Key components of this tasks are the interplay between needle holder and the surgical needle. Reliable 3D localization of needle and instruments in real time could be used to augment the scene with additional parameters that describe their quantitative geometric relation, e.g. the relation between the estimated needle plane and its rotation center and the instrument. This could contribute towards standardization and training of basic skills and operative techniques, enhance overall surgical performance, and reduce the risk of complications. The paper proposes an Augmented Reality environment with quantitative and qualitative visual representations to enhance laparoscopic training outcomes performed on a silicone pad. This is enabled by a multi-task supervised deep neural network which performs multi-class segmentation and depth map prediction. Scarcity of labels has been conquered by creating a virtual environment which resembles the surgical training scenario to generate dense depth maps and segmentation maps. The proposed convolutional neural network was tested on real surgical training scenarios and showed to be robust to occlusion of the needle. The network achieves a dice score of 0.67 for surgical needle segmentation, 0.81 for needle holder instrument segmentation and a mean absolute error of 6.5 mm for depth estimation.
Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation
Jul 05, 2019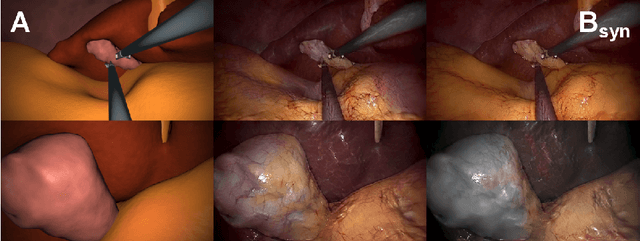
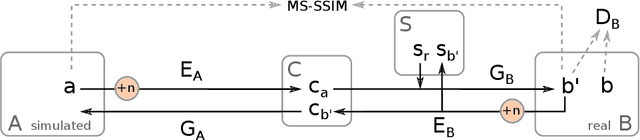
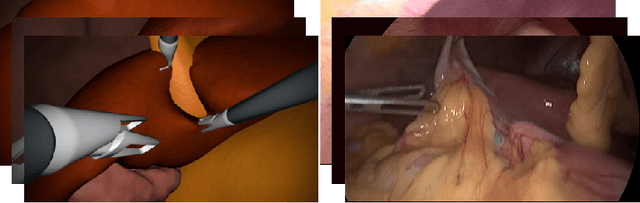
Abstract:In the medical domain, the lack of large training data sets and benchmarks is often a limiting factor for training deep neural networks. In contrast to expensive manual labeling, computer simulations can generate large and fully labeled data sets with a minimum of manual effort. However, models that are trained on simulated data usually do not translate well to real scenarios. To bridge the domain gap between simulated and real laparoscopic images, we exploit recent advances in unpaired image-to-image translation. We extent an image-to-image translation method to generate a diverse multitude of realistically looking synthetic images based on images from a simple laparoscopy simulation. By incorporating means to ensure that the image content is preserved during the translation process, we ensure that the labels given for the simulated images remain valid for their realistically looking translations. This way, we are able to generate a large, fully labeled synthetic data set of laparoscopic images with realistic appearance. We show that this data set can be used to train models for the task of liver segmentation of laparoscopic images. We achieve average dice scores of up to 0.89 in some patients without manually labeling a single laparoscopic image and show that using our synthetic data to pre-train models can greatly improve their performance. The synthetic data set will be made publicly available, fully labeled with segmentation maps, depth maps, normal maps, and positions of tools and camera (http://opencas.dkfz.de/image2image).
Cross-Domain Conditional Generative Adversarial Networks for Stereoscopic Hyperrealism in Surgical Training
Jun 24, 2019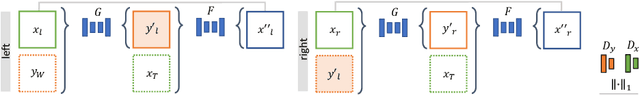
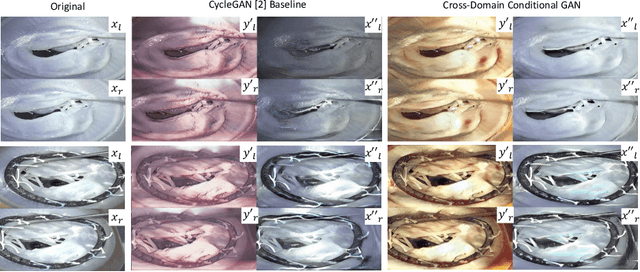
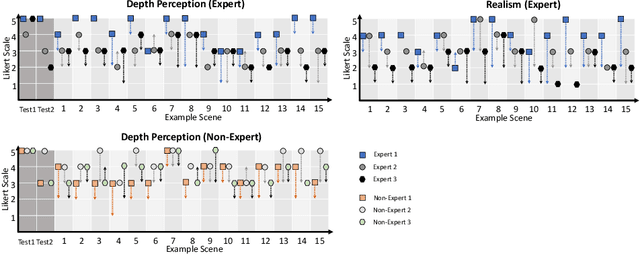
Abstract:Phantoms for surgical training are able to mimic cutting and suturing properties and patient-individual shape of organs, but lack a realistic visual appearance that captures the heterogeneity of surgical scenes. In order to overcome this in endoscopic approaches, hyperrealistic concepts have been proposed to be used in an augmented reality-setting, which are based on deep image-to-image transformation methods. Such concepts are able to generate realistic representations of phantoms learned from real intraoperative endoscopic sequences. Conditioned on frames from the surgical training process, the learned models are able to generate impressive results by transforming unrealistic parts of the image (e.g.\ the uniform phantom texture is replaced by the more heterogeneous texture of the tissue). Image-to-image synthesis usually learns a mapping $G:X~\to~Y$ such that the distribution of images from $G(X)$ is indistinguishable from the distribution $Y$. However, it does not necessarily force the generated images to be consistent and without artifacts. In the endoscopic image domain this can affect depth cues and stereo consistency of a stereo image pair, which ultimately impairs surgical vision. We propose a cross-domain conditional generative adversarial network approach (GAN) that aims to generate more consistent stereo pairs. The results show substantial improvements in depth perception and realism evaluated by 3 domain experts and 3 medical students on a 3D monitor over the baseline method. In 84 of 90 instances our proposed method was preferred or rated equal to the baseline.
Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features
Jan 25, 2018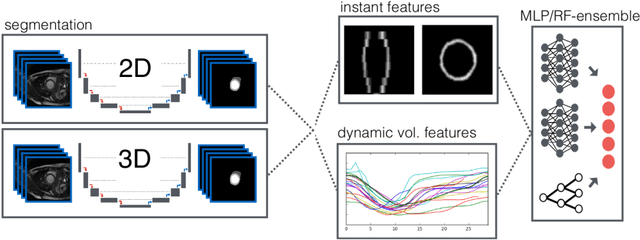
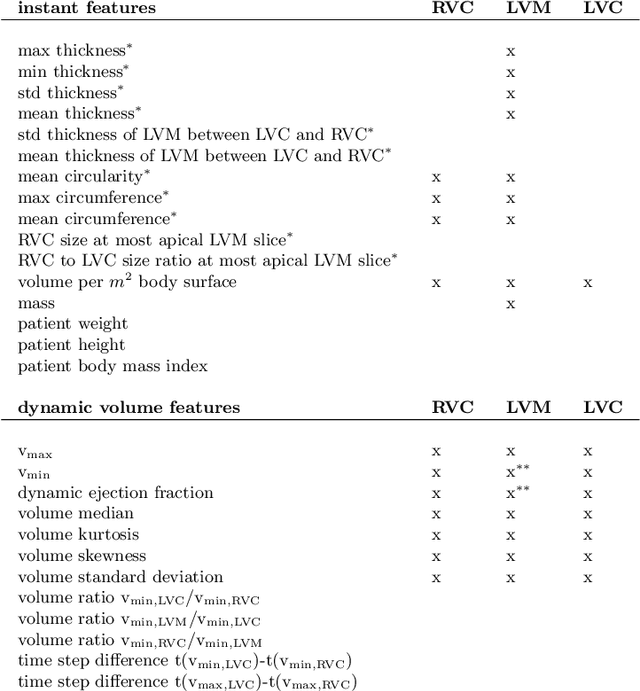
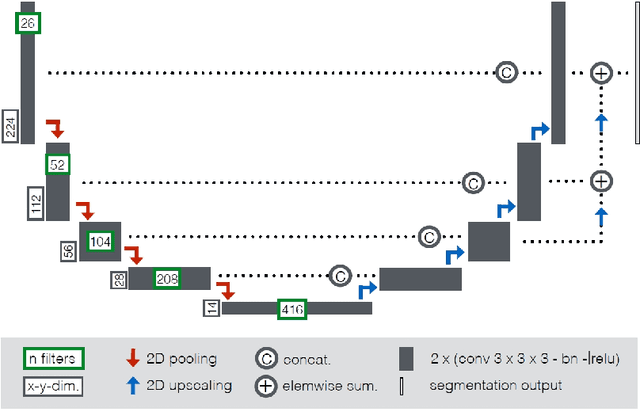
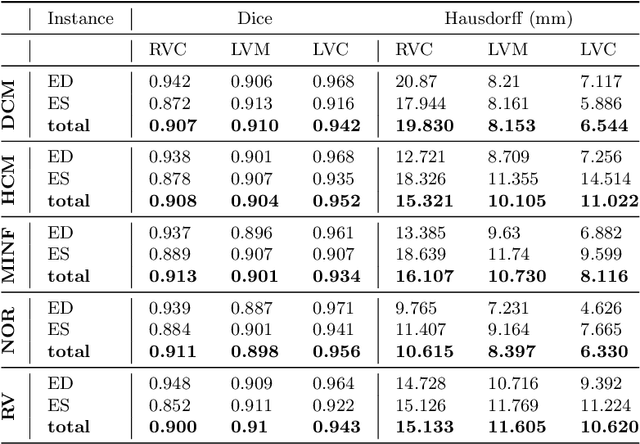
Abstract:Cardiac magnetic resonance imaging improves on diagnosis of cardiovascular diseases by providing images at high spatiotemporal resolution. Manual evaluation of these time-series, however, is expensive and prone to biased and non-reproducible outcomes. In this paper, we present a method that addresses named limitations by integrating segmentation and disease classification into a fully automatic processing pipeline. We use an ensemble of UNet inspired architectures for segmentation of cardiac structures such as the left and right ventricular cavity (LVC, RVC) and the left ventricular myocardium (LVM) on each time instance of the cardiac cycle. For the classification task, information is extracted from the segmented time-series in form of comprehensive features handcrafted to reflect diagnostic clinical procedures. Based on these features we train an ensemble of heavily regularized multilayer perceptrons (MLP) and a random forest classifier to predict the pathologic target class. We evaluated our method on the ACDC dataset (4 pathology groups, 1 healthy group) and achieve dice scores of 0.945 (LVC), 0.908 (RVC) and 0.905 (LVM) in a cross-validation over the training set (100 cases) and 0.950 (LVC), 0.923 (RVC) and 0.911 (LVM) on the test set (50 cases). We report a classification accuracy of 94% on a training set cross-validation and 92% on the test set. Our results underpin the potential of machine learning methods for accurate, fast and reproducible segmentation and computer-assisted diagnosis (CAD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge