Maria R. Robu
More unlabelled data or label more data? A study on semi-supervised laparoscopic image segmentation
Aug 20, 2019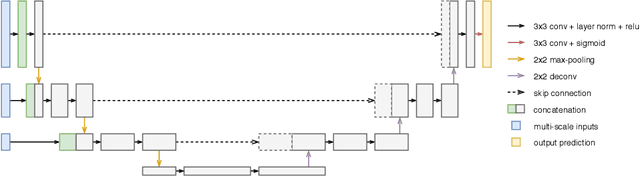
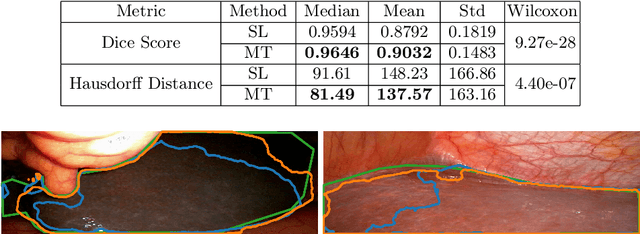
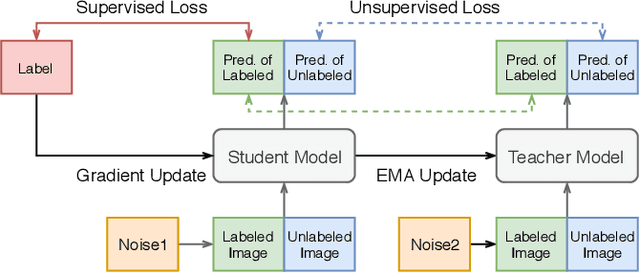
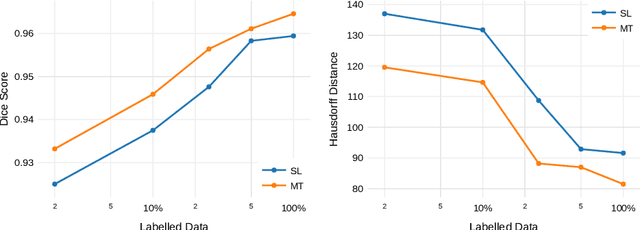
Abstract:Improving a semi-supervised image segmentation task has the option of adding more unlabelled images, labelling the unlabelled images or combining both, as neither image acquisition nor expert labelling can be considered trivial in most clinical applications. With a laparoscopic liver image segmentation application, we investigate the performance impact by altering the quantities of labelled and unlabelled training data, using a semi-supervised segmentation algorithm based on the mean teacher learning paradigm. We first report a significantly higher segmentation accuracy, compared with supervised learning. Interestingly, this comparison reveals that the training strategy adopted in the semi-supervised algorithm is also responsible for this observed improvement, in addition to the added unlabelled data. We then compare different combinations of labelled and unlabelled data set sizes for training semi-supervised segmentation networks, to provide a quantitative example of the practically useful trade-off between the two data planning strategies in this surgical guidance application.
Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation
Jul 05, 2019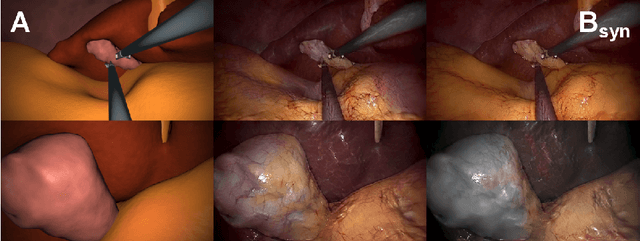
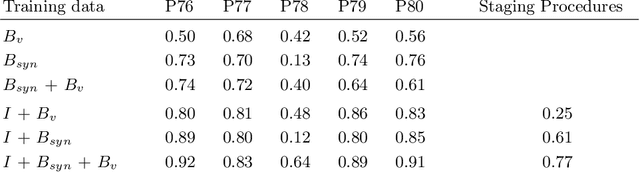
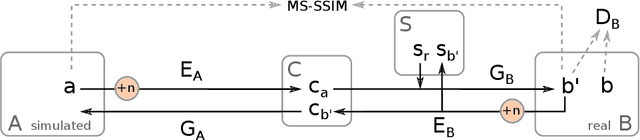
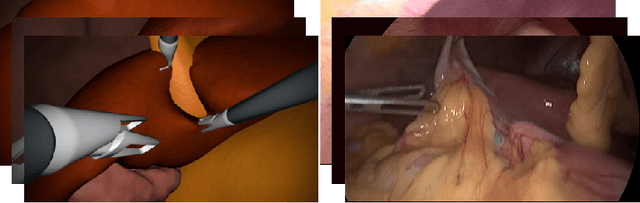
Abstract:In the medical domain, the lack of large training data sets and benchmarks is often a limiting factor for training deep neural networks. In contrast to expensive manual labeling, computer simulations can generate large and fully labeled data sets with a minimum of manual effort. However, models that are trained on simulated data usually do not translate well to real scenarios. To bridge the domain gap between simulated and real laparoscopic images, we exploit recent advances in unpaired image-to-image translation. We extent an image-to-image translation method to generate a diverse multitude of realistically looking synthetic images based on images from a simple laparoscopy simulation. By incorporating means to ensure that the image content is preserved during the translation process, we ensure that the labels given for the simulated images remain valid for their realistically looking translations. This way, we are able to generate a large, fully labeled synthetic data set of laparoscopic images with realistic appearance. We show that this data set can be used to train models for the task of liver segmentation of laparoscopic images. We achieve average dice scores of up to 0.89 in some patients without manually labeling a single laparoscopic image and show that using our synthetic data to pre-train models can greatly improve their performance. The synthetic data set will be made publicly available, fully labeled with segmentation maps, depth maps, normal maps, and positions of tools and camera (http://opencas.dkfz.de/image2image).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge