Rishabh Anand
Zatom-1: A Multimodal Flow Foundation Model for 3D Molecules and Materials
Feb 24, 2026Abstract:General-purpose 3D chemical modeling encompasses molecules and materials, requiring both generative and predictive capabilities. However, most existing AI approaches are optimized for a single domain (molecules or materials) and a single task (generation or prediction), which limits representation sharing and transfer. We introduce Zatom-1, the first foundation model that unifies generative and predictive learning of 3D molecules and materials. Zatom-1 is a Transformer trained with a multimodal flow matching objective that jointly models discrete atom types and continuous 3D geometries. This approach supports scalable pretraining with predictable gains as model capacity increases, while enabling fast and stable sampling. We use joint generative pretraining as a universal initialization for downstream multi-task prediction of properties, energies, and forces. Empirically, Zatom-1 matches or outperforms specialized baselines on both generative and predictive benchmarks, while reducing the generative inference time by more than an order of magnitude. Our experiments demonstrate positive predictive transfer between chemical domains from joint generative pretraining: modeling materials during pretraining improves molecular property prediction accuracy.
HELM: Hyperbolic Large Language Models via Mixture-of-Curvature Experts
May 30, 2025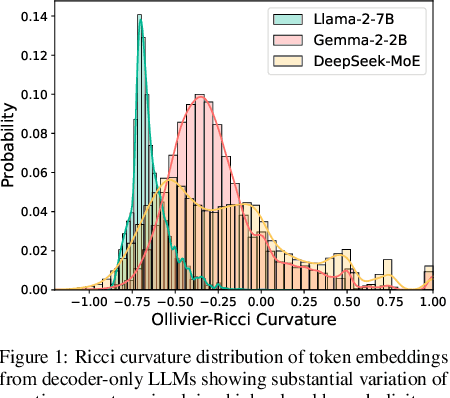
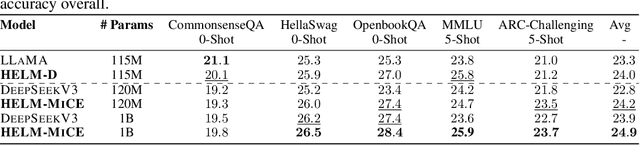
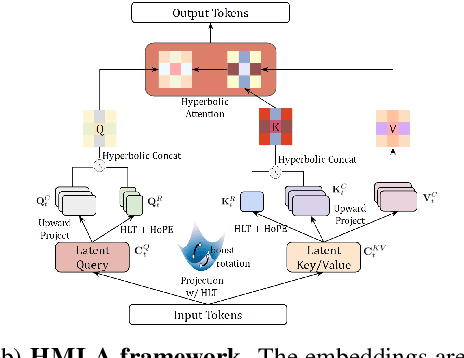

Abstract:Large language models (LLMs) have shown great success in text modeling tasks across domains. However, natural language exhibits inherent semantic hierarchies and nuanced geometric structure, which current LLMs do not capture completely owing to their reliance on Euclidean operations. Recent studies have also shown that not respecting the geometry of token embeddings leads to training instabilities and degradation of generative capabilities. These findings suggest that shifting to non-Euclidean geometries can better align language models with the underlying geometry of text. We thus propose to operate fully in Hyperbolic space, known for its expansive, scale-free, and low-distortion properties. We thus introduce HELM, a family of HypErbolic Large Language Models, offering a geometric rethinking of the Transformer-based LLM that addresses the representational inflexibility, missing set of necessary operations, and poor scalability of existing hyperbolic LMs. We additionally introduce a Mixture-of-Curvature Experts model, HELM-MICE, where each expert operates in a distinct curvature space to encode more fine-grained geometric structure from text, as well as a dense model, HELM-D. For HELM-MICE, we further develop hyperbolic Multi-Head Latent Attention (HMLA) for efficient, reduced-KV-cache training and inference. For both models, we develop essential hyperbolic equivalents of rotary positional encodings and RMS normalization. We are the first to train fully hyperbolic LLMs at billion-parameter scale, and evaluate them on well-known benchmarks such as MMLU and ARC, spanning STEM problem-solving, general knowledge, and commonsense reasoning. Our results show consistent gains from our HELM architectures -- up to 4% -- over popular Euclidean architectures used in LLaMA and DeepSeek, highlighting the efficacy and enhanced reasoning afforded by hyperbolic geometry in large-scale LM pretraining.
RNA-FrameFlow: Flow Matching for de novo 3D RNA Backbone Design
Jun 19, 2024Abstract:We introduce RNA-FrameFlow, the first generative model for 3D RNA backbone design. We build upon SE(3) flow matching for protein backbone generation and establish protocols for data preparation and evaluation to address unique challenges posed by RNA modeling. We formulate RNA structures as a set of rigid-body frames and associated loss functions which account for larger, more conformationally flexible RNA backbones (13 atoms per nucleotide) vs. proteins (4 atoms per residue). Toward tackling the lack of diversity in 3D RNA datasets, we explore training with structural clustering and cropping augmentations. Additionally, we define a suite of evaluation metrics to measure whether the generated RNA structures are globally self-consistent (via inverse folding followed by forward folding) and locally recover RNA-specific structural descriptors. The most performant version of RNA-FrameFlow generates locally realistic RNA backbones of 40-150 nucleotides, over 40% of which pass our validity criteria as measured by a self-consistency TM-score >= 0.45, at which two RNAs have the same global fold. Open-source code: https://github.com/rish-16/rna-backbone-design
Pretrained Encoders are All You Need
Jun 09, 2021



Abstract:Data-efficiency and generalization are key challenges in deep learning and deep reinforcement learning as many models are trained on large-scale, domain-specific, and expensive-to-label datasets. Self-supervised models trained on large-scale uncurated datasets have shown successful transfer to diverse settings. We investigate using pretrained image representations and spatio-temporal attention for state representation learning in Atari. We also explore fine-tuning pretrained representations with self-supervised techniques, i.e., contrastive predictive coding, spatio-temporal contrastive learning, and augmentations. Our results show that pretrained representations are at par with state-of-the-art self-supervised methods trained on domain-specific data. Pretrained representations, thus, yield data and compute-efficient state representations. https://github.com/PAL-ML/PEARL_v1
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge