Pengbo Liu
Multi-site Organ Segmentation with Federated Partial Supervision and Site Adaptation
Feb 08, 2023



Abstract:Objective and Impact Statement: Accurate organ segmentation is critical for many clinical applications at different clinical sites, which may have their specific application requirements that concern different organs. Introduction: However, learning high-quality, site-specific organ segmentation models is challenging as it often needs on-site curation of a large number of annotated images. Security concerns further complicate the matter. Methods: The paper aims to tackle these challenges via a two-phase aggregation-then-adaptation approach. The first phase of federated aggregation learns a single multi-organ segmentation model by leveraging the strength of 'bigger data', which are formed by (i) aggregating together datasets from multiple sites that with different organ labels to provide partial supervision, and (ii) conducting partially supervised learning without data breach. The second phase of site adaptation is to transfer the federated multi-organ segmentation model to site-specific organ segmentation models, one model per site, in order to further improve the performance of each site's organ segmentation task. Furthermore, improved marginal loss and exclusion loss functions are used to avoid 'knowledge conflict' problem in a partially supervision mechanism. Results and Conclusion: Extensive experiments on five organ segmentation datasets demonstrate the effectiveness of our multi-site approach, significantly outperforming the site-per-se learned models and achieving the performance comparable to the centrally learned models.
Learning Incrementally to Segment Multiple Organs in a CT Image
Mar 04, 2022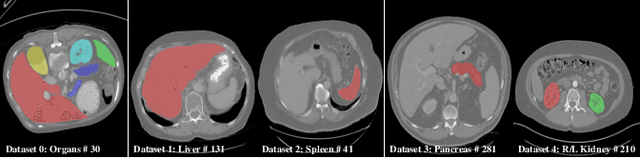
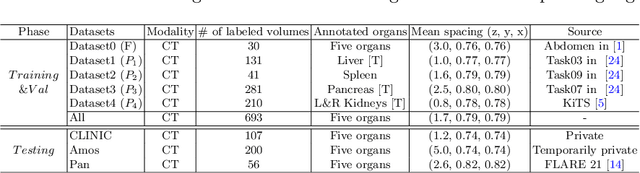
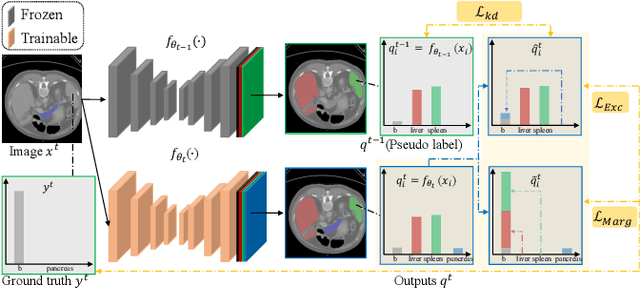
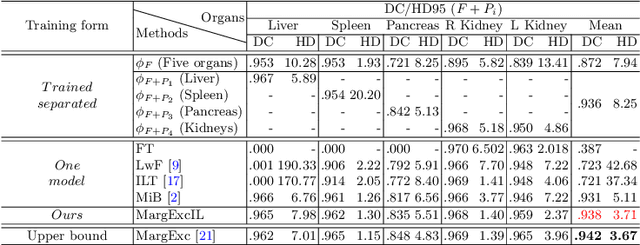
Abstract:There exists a large number of datasets for organ segmentation, which are partially annotated and sequentially constructed. A typical dataset is constructed at a certain time by curating medical images and annotating the organs of interest. In other words, new datasets with annotations of new organ categories are built over time. To unleash the potential behind these partially labeled, sequentially-constructed datasets, we propose to incrementally learn a multi-organ segmentation model. In each incremental learning (IL) stage, we lose the access to previous data and annotations, whose knowledge is assumingly captured by the current model, and gain the access to a new dataset with annotations of new organ categories, from which we learn to update the organ segmentation model to include the new organs. While IL is notorious for its `catastrophic forgetting' weakness in the context of natural image analysis, we experimentally discover that such a weakness mostly disappears for CT multi-organ segmentation. To further stabilize the model performance across the IL stages, we introduce a light memory module and some loss functions to restrain the representation of different categories in feature space, aggregating feature representation of the same class and separating feature representation of different classes. Extensive experiments on five open-sourced datasets are conducted to illustrate the effectiveness of our method.
Universal Segmentation of 33 Anatomies
Mar 04, 2022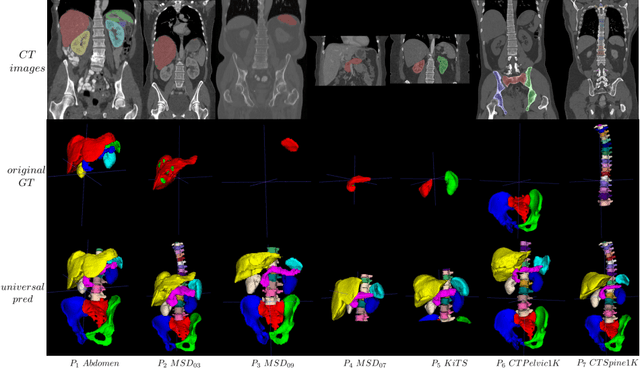
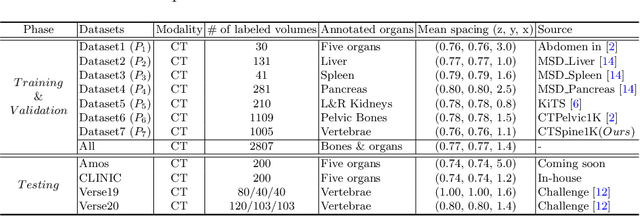
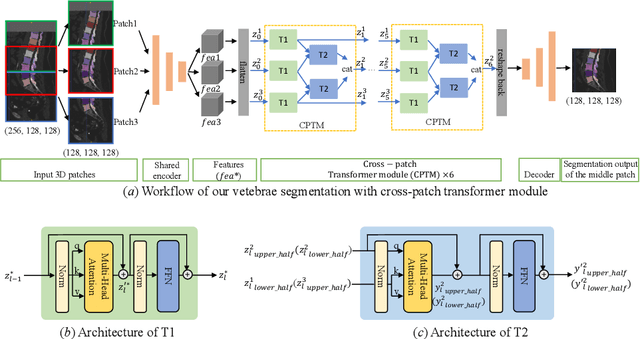
Abstract:In the paper, we present an approach for learning a single model that universally segments 33 anatomical structures, including vertebrae, pelvic bones, and abdominal organs. Our model building has to address the following challenges. Firstly, while it is ideal to learn such a model from a large-scale, fully-annotated dataset, it is practically hard to curate such a dataset. Thus, we resort to learn from a union of multiple datasets, with each dataset containing the images that are partially labeled. Secondly, along the line of partial labelling, we contribute an open-source, large-scale vertebra segmentation dataset for the benefit of spine analysis community, CTSpine1K, boasting over 1,000 3D volumes and over 11K annotated vertebrae. Thirdly, in a 3D medical image segmentation task, due to the limitation of GPU memory, we always train a model using cropped patches as inputs instead a whole 3D volume, which limits the amount of contextual information to be learned. To this, we propose a cross-patch transformer module to fuse more information in adjacent patches, which enlarges the aggregated receptive field for improved segmentation performance. This is especially important for segmenting, say, the elongated spine. Based on 7 partially labeled datasets that collectively contain about 2,800 3D volumes, we successfully learn such a universal model. Finally, we evaluate the universal model on multiple open-source datasets, proving that our model has a good generalization performance and can potentially serve as a solid foundation for downstream tasks.
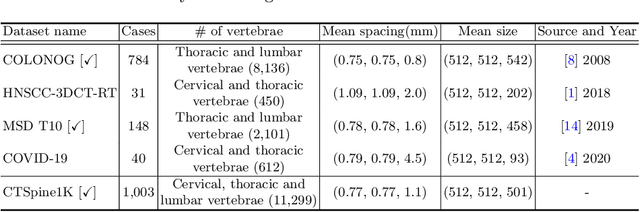
CTSpine1K: A Large-Scale Dataset for Spinal Vertebrae Segmentation in Computed Tomography
Jun 10, 2021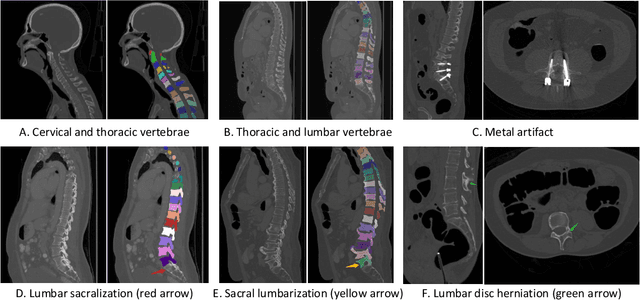
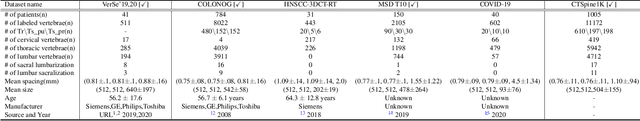
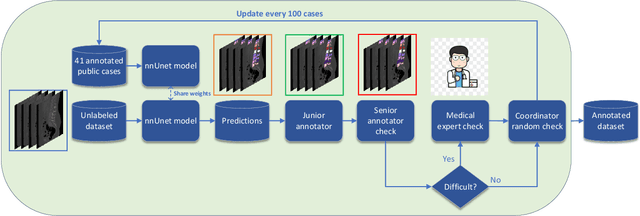
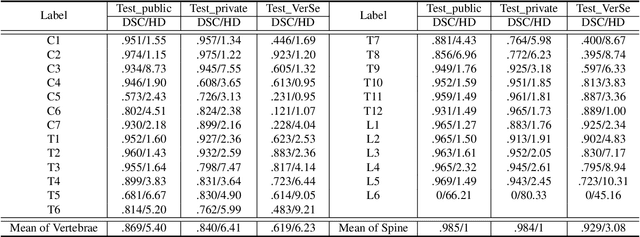
Abstract:Spine-related diseases have high morbidity and cause a huge burden of social cost. Spine imaging is an essential tool for noninvasively visualizing and assessing spinal pathology. Segmenting vertebrae in computed tomography (CT) images is the basis of quantitative medical image analysis for clinical diagnosis and surgery planning of spine diseases. Current publicly available annotated datasets on spinal vertebrae are small in size. Due to the lack of a large-scale annotated spine image dataset, the mainstream deep learning-based segmentation methods, which are data-driven, are heavily restricted. In this paper, we introduce a large-scale spine CT dataset, called CTSpine1K, curated from multiple sources for vertebra segmentation, which contains 1,005 CT volumes with over 11,100 labeled vertebrae belonging to different spinal conditions. Based on this dataset, we conduct several spinal vertebrae segmentation experiments to set the first benchmark. We believe that this large-scale dataset will facilitate further research in many spine-related image analysis tasks, including but not limited to vertebrae segmentation, labeling, 3D spine reconstruction from biplanar radiographs, image super-resolution, and enhancement.
Gumbel-Attention for Multi-modal Machine Translation
Mar 16, 2021
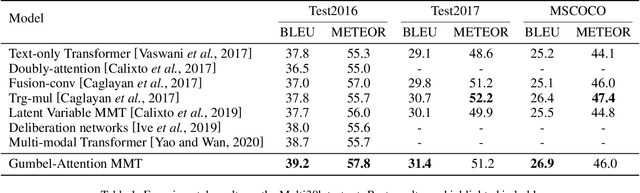
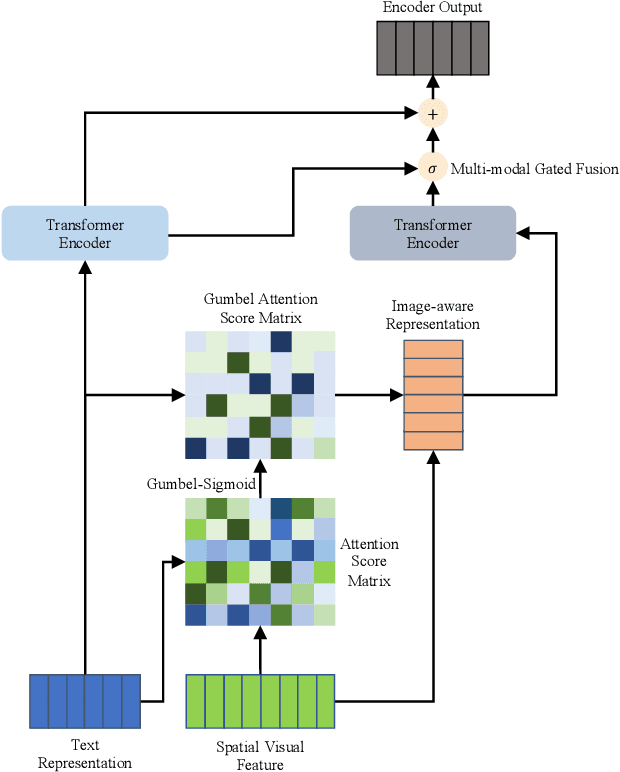
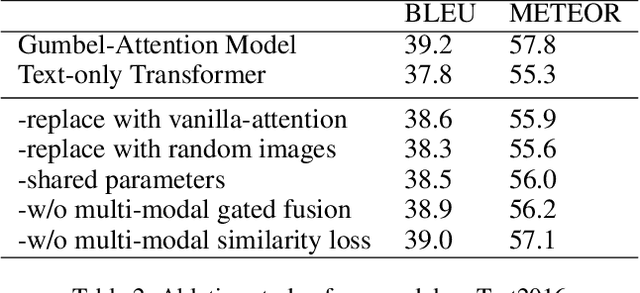
Abstract:Multi-modal machine translation (MMT) improves translation quality by introducing visual information. However, the existing MMT model ignores the problem that the image will bring information irrelevant to the text, causing much noise to the model and affecting the translation quality. In this paper, we propose a novel Gumbel-Attention for multi-modal machine translation, which selects the text-related parts of the image features. Specifically, different from the previous attention-based method, we first use a differentiable method to select the image information and automatically remove the useless parts of the image features. Through the score matrix of Gumbel-Attention and image features, the image-aware text representation is generated. And then, we independently encode the text representation and the image-aware text representation with the multi-modal encoder. Finally, the final output of the encoder is obtained through multi-modal gated fusion. Experiments and case analysis proves that our method retains the image features related to the text, and the remaining parts help the MMT model generates better translations.
Incremental Learning for Multi-organ Segmentation with Partially Labeled Datasets
Mar 08, 2021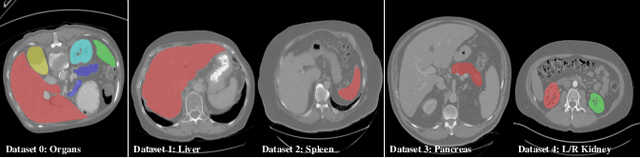
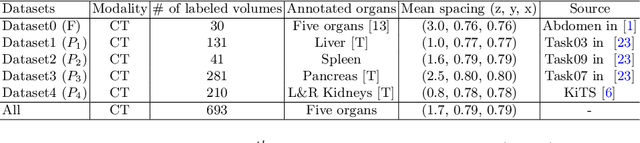
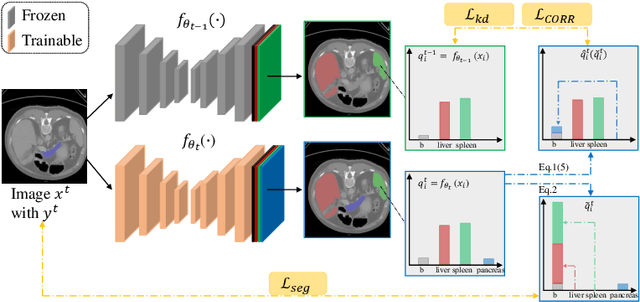
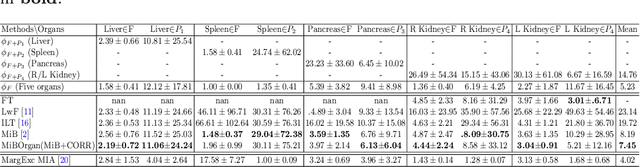
Abstract:There exists a large number of datasets for organ segmentation, which are partially annotated, and sequentially constructed. A typical dataset is constructed at a certain time by curating medical images and annotating the organs of interest. In other words, new datasets with annotations of new organ categories are built over time. To unleash the potential behind these partially labeled, sequentially-constructed datasets, we propose to learn a multi-organ segmentation model through incremental learning (IL). In each IL stage, we lose access to the previous annotations, whose knowledge is assumingly captured by the current model, and gain the access to a new dataset with annotations of new organ categories, from which we learn to update the organ segmentation model to include the new organs. We give the first attempt to conjecture that the different distribution is the key reason for 'catastrophic forgetting' that commonly exists in IL methods, and verify that IL has the natural adaptability to medical image scenarios. Extensive experiments on five open-sourced datasets are conducted to prove the effectiveness of our method and the conjecture mentioned above.
Deep Learning to Segment Pelvic Bones: Large-scale CT Datasets and Baseline Models
Dec 16, 2020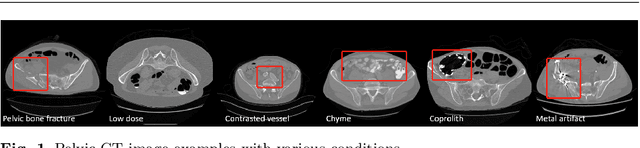
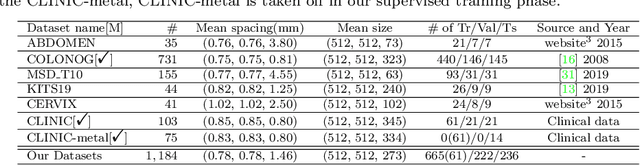
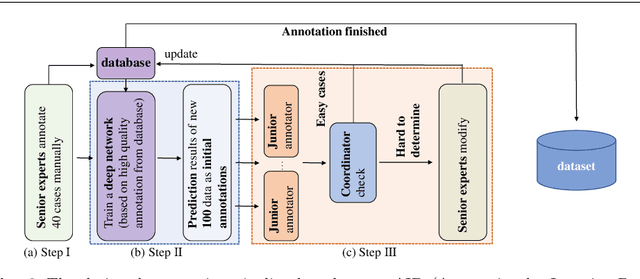
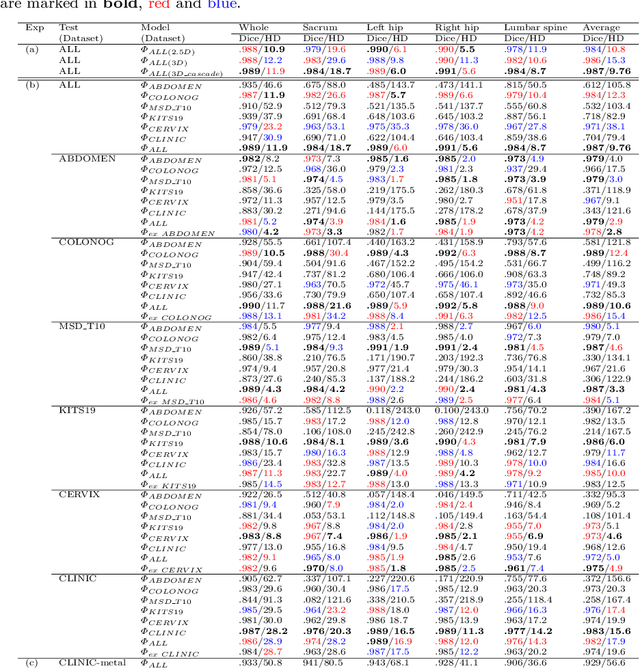
Abstract:Purpose: Pelvic bone segmentation in CT has always been an essential step in clinical diagnosis and surgery planning of pelvic bone diseases. Existing methods for pelvic bone segmentation are either hand-crafted or semi-automatic and achieve limited accuracy when dealing with image appearance variations due to the multi-site domain shift, the presence of contrasted vessels, coprolith and chyme, bone fractures, low dose, metal artifacts, etc. Due to the lack of a large-scale pelvic CT dataset with annotations, deep learning methods are not fully explored. Methods: In this paper, we aim to bridge the data gap by curating a large pelvic CT dataset pooled from multiple sources and different manufacturers, including 1, 184 CT volumes and over 320, 000 slices with different resolutions and a variety of the above-mentioned appearance variations. Then we propose for the first time, to the best of our knowledge, to learn a deep multi-class network for segmenting lumbar spine, sacrum, left hip, and right hip, from multiple-domain images simultaneously to obtain more effective and robust feature representations. Finally, we introduce a post-processing tool based on the signed distance function (SDF) to eliminate false predictions while retaining correctly predicted bone fragments. Results: Extensive experiments on our dataset demonstrate the effectiveness of our automatic method, achieving an average Dice of 0.987 for a metal-free volume. SDF post-processor yields a decrease of 10.5% in hausdorff distance by maintaining important bone fragments in post-processing phase. Conclusion: We believe this large-scale dataset will promote the development of the whole community and plan to open source the images, annotations, codes, and trained baseline models at this URL1.
Exploiting Typed Syntactic Dependencies for Targeted Sentiment Classification Using Graph Attention Neural Network
Feb 22, 2020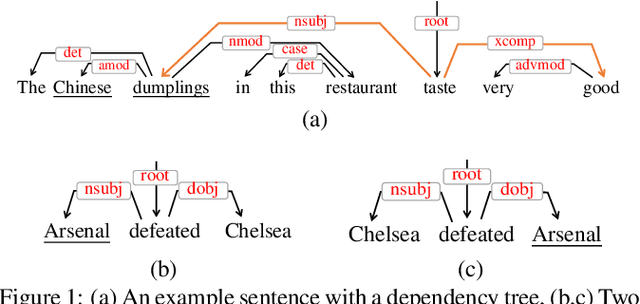
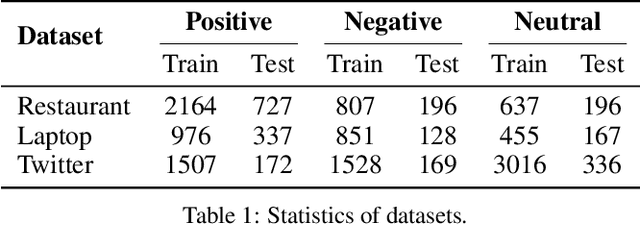
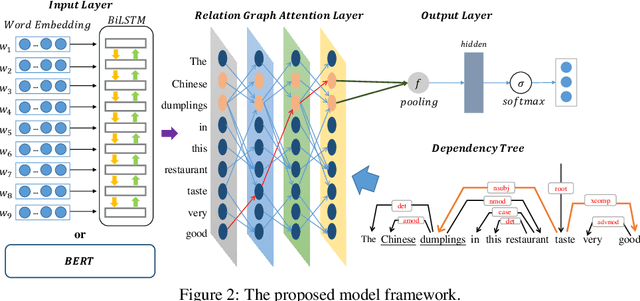
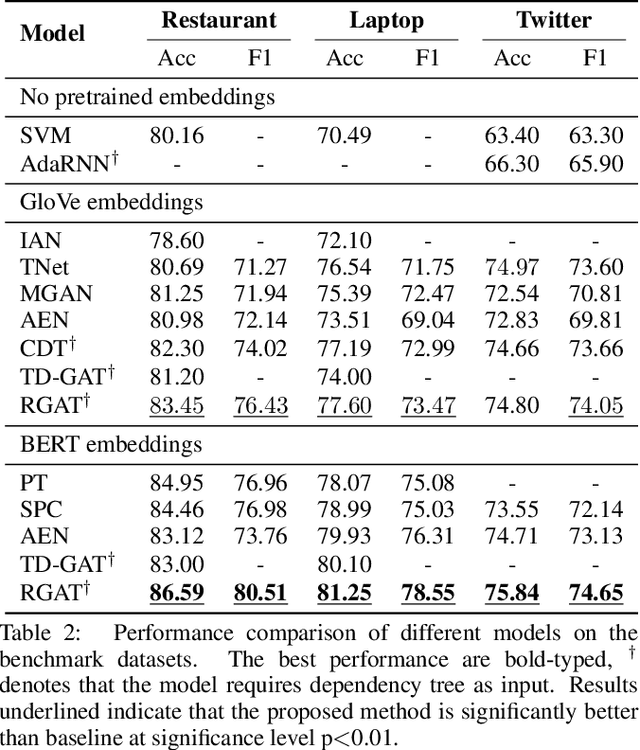
Abstract:Targeted sentiment classification predicts the sentiment polarity on given target mentions in input texts. Dominant methods employ neural networks for encoding the input sentence and extracting relations between target mentions and their contexts. Recently, graph neural network has been investigated for integrating dependency syntax for the task, achieving the state-of-the-art results. However, existing methods do not consider dependency label information, which can be intuitively useful. To solve the problem, we investigate a novel relational graph attention network that integrates typed syntactic dependency information. Results on standard benchmarks show that our method can effectively leverage label information for improving targeted sentiment classification performances. Our final model significantly outperforms state-of-the-art syntax-based approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge