Michael S. Yao
Diversity By Design: Leveraging Distribution Matching for Offline Model-Based Optimization
Jan 30, 2025

Abstract:The goal of offline model-based optimization (MBO) is to propose new designs that maximize a reward function given only an offline dataset. However, an important desiderata is to also propose a diverse set of final candidates that capture many optimal and near-optimal design configurations. We propose Diversity in Adversarial Model-based Optimization (DynAMO) as a novel method to introduce design diversity as an explicit objective into any MBO problem. Our key insight is to formulate diversity as a distribution matching problem where the distribution of generated designs captures the inherent diversity contained within the offline dataset. Extensive experiments spanning multiple scientific domains show that DynAMO can be used with common optimization methods to significantly improve the diversity of proposed designs while still discovering high-quality candidates.
Evidence Is All You Need: Ordering Imaging Studies via Language Model Alignment with the ACR Appropriateness Criteria
Sep 27, 2024



Abstract:Diagnostic imaging studies are an increasingly important component of the workup and management of acutely presenting patients. However, ordering appropriate imaging studies according to evidence-based medical guidelines is a challenging task with a high degree of variability between healthcare providers. To address this issue, recent work has investigated if generative AI and large language models can be leveraged to help clinicians order relevant imaging studies for patients. However, it is challenging to ensure that these tools are correctly aligned with medical guidelines, such as the American College of Radiology's Appropriateness Criteria (ACR AC). In this study, we introduce a framework to intelligently leverage language models by recommending imaging studies for patient cases that are aligned with evidence-based guidelines. We make available a novel dataset of patient "one-liner" scenarios to power our experiments, and optimize state-of-the-art language models to achieve an accuracy on par with clinicians in image ordering. Finally, we demonstrate that our language model-based pipeline can be used as intelligent assistants by clinicians to support image ordering workflows and improve the accuracy of imaging study ordering according to the ACR AC. Our work demonstrates and validates a strategy to leverage AI-based software to improve trustworthy clinical decision making in alignment with expert evidence-based guidelines.
A Textbook Remedy for Domain Shifts: Knowledge Priors for Medical Image Analysis
May 23, 2024



Abstract:While deep networks have achieved broad success in analyzing natural images, when applied to medical scans, they often fail in unexcepted situations. We investigate this challenge and focus on model sensitivity to domain shifts, such as data sampled from different hospitals or data confounded by demographic variables such as sex, race, etc, in the context of chest X-rays and skin lesion images. A key finding we show empirically is that existing visual backbones lack an appropriate prior from the architecture for reliable generalization in these settings. Taking inspiration from medical training, we propose giving deep networks a prior grounded in explicit medical knowledge communicated in natural language. To this end, we introduce Knowledge-enhanced Bottlenecks (KnoBo), a class of concept bottleneck models that incorporates knowledge priors that constrain it to reason with clinically relevant factors found in medical textbooks or PubMed. KnoBo uses retrieval-augmented language models to design an appropriate concept space paired with an automatic training procedure for recognizing the concept. We evaluate different resources of knowledge and recognition architectures on a broad range of domain shifts across 20 datasets. In our comprehensive evaluation with two imaging modalities, KnoBo outperforms fine-tuned models on confounded datasets by 32.4% on average. Finally, evaluations reveal that PubMed is a promising resource for making medical models less sensitive to domain shift, outperforming other resources on both diversity of information and final prediction performance.
A Concept-based Interpretable Model for the Diagnosis of Choroid Neoplasias using Multimodal Data
Mar 08, 2024Abstract:Diagnosing rare diseases presents a common challenge in clinical practice, necessitating the expertise of specialists for accurate identification. The advent of machine learning offers a promising solution, while the development of such technologies is hindered by the scarcity of data on rare conditions and the demand for models that are both interpretable and trustworthy in a clinical context. Interpretable AI, with its capacity for human-readable outputs, can facilitate validation by clinicians and contribute to medical education. In the current work, we focus on choroid neoplasias, the most prevalent form of eye cancer in adults, albeit rare with 5.1 per million. We built the so-far largest dataset consisting of 750 patients, incorporating three distinct imaging modalities collected from 2004 to 2022. Our work introduces a concept-based interpretable model that distinguishes between three types of choroidal tumors, integrating insights from domain experts via radiological reports. Remarkably, this model not only achieves an F1 score of 0.91, rivaling that of black-box models, but also boosts the diagnostic accuracy of junior doctors by 42%. This study highlights the significant potential of interpretable machine learning in improving the diagnosis of rare diseases, laying a groundwork for future breakthroughs in medical AI that could tackle a wider array of complex health scenarios.
Generative Adversarial Bayesian Optimization for Surrogate Objectives
Feb 09, 2024Abstract:Offline model-based policy optimization seeks to optimize a learned surrogate objective function without querying the true oracle objective during optimization. However, inaccurate surrogate model predictions are frequently encountered along the optimization trajectory. To address this limitation, we propose generative adversarial Bayesian optimization (GABO) using adaptive source critic regularization, a task-agnostic framework for Bayesian optimization that employs a Lipschitz-bounded source critic model to constrain the optimization trajectory to regions where the surrogate function is reliable. We show that under certain assumptions for the continuous input space prior, our algorithm dynamically adjusts the strength of the source critic regularization. GABO outperforms existing baselines on a number of different offline optimization tasks across a variety of scientific domains. Our code is available at https://github.com/michael-s-yao/gabo
Learning-Based Radiomic Prediction of Type 2 Diabetes Mellitus Using Image-Derived Phenotypes
Sep 20, 2022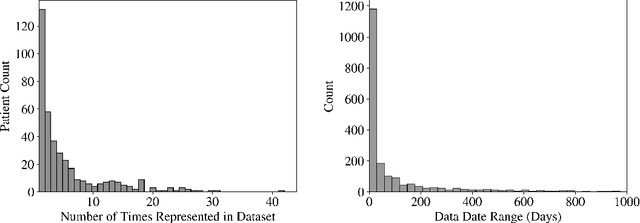
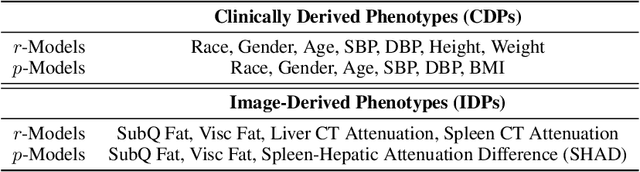
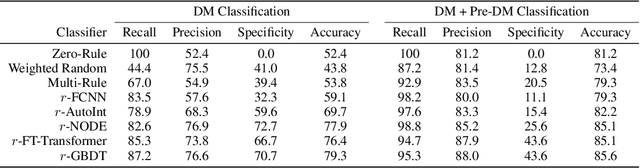
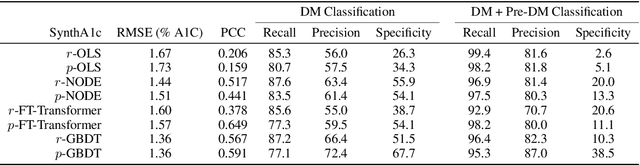
Abstract:Early diagnosis of Type 2 Diabetes Mellitus (T2DM) is crucial to enable timely therapeutic interventions and lifestyle modifications. As medical imaging data become more widely available for many patient populations, we sought to investigate whether image-derived phenotypic data could be leveraged in tabular learning classifier models to predict T2DM incidence without the use of invasive blood lab measurements. We show that both neural network and decision tree models that use image-derived phenotypes can predict patient T2DM status with recall scores as high as 87.6%. We also propose the novel use of these same architectures as 'SynthA1c encoders' that are able to output interpretable values mimicking blood hemoglobin A1C empirical lab measurements. Finally, we demonstrate that T2DM risk prediction model sensitivity to small perturbations in input vector components can be used to predict performance on covariates sampled from previously unseen patient populations.
A Path Towards Clinical Adaptation of Accelerated MRI
Aug 26, 2022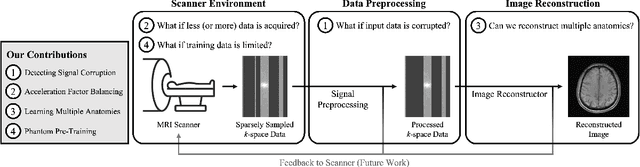
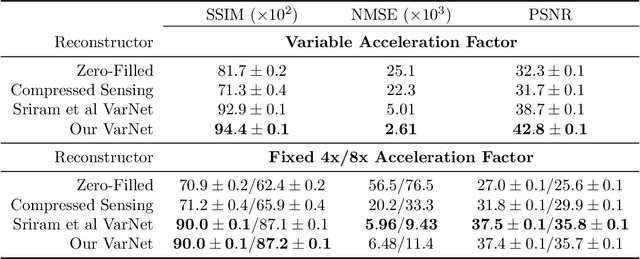
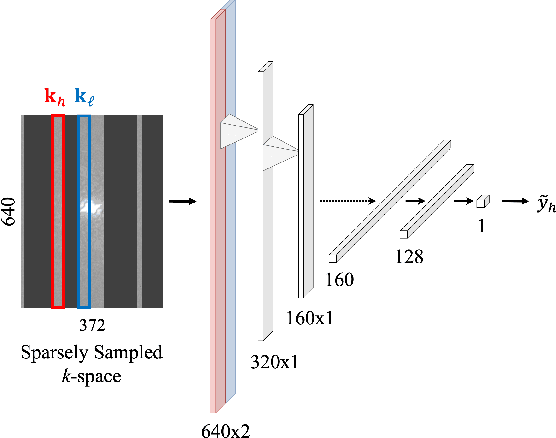
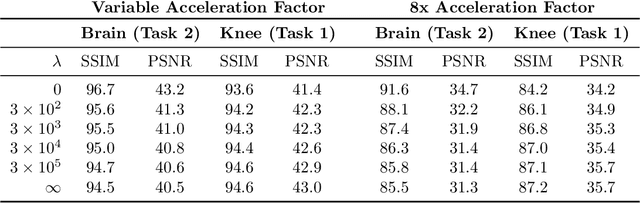
Abstract:Accelerated MRI reconstructs images of clinical anatomies from sparsely sampled signal data to reduce patient scan times. While recent works have leveraged deep learning to accomplish this task, such approaches have often only been explored in simulated environments where there is no signal corruption or resource limitations. In this work, we explore augmentations to neural network MRI image reconstructors to enhance their clinical relevancy. Namely, we propose a ConvNet model for detecting sources of image artifacts that achieves a classifer $F_2$ score of $79.1\%$. We also demonstrate that training reconstructors on MR signal data with variable acceleration factors can improve their average performance during a clinical patient scan by up to $2\%$. We offer a loss function to overcome catastrophic forgetting when models learn to reconstruct MR images of multiple anatomies and orientations. Finally, we propose a method for using simulated phantom data to pre-train reconstructors in situations with limited clinically acquired datasets and compute capabilities. Our results provide a potential path forward for clinical adaptation of accelerated MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge