Drew A. Torigian
RSNA Large Language Model Benchmark Dataset for Chest Radiographs of Cardiothoracic Disease: Radiologist Evaluation and Validation Enhanced by AI Labels (REVEAL-CXR)
Jan 21, 2026Abstract:Multimodal large language models have demonstrated comparable performance to that of radiology trainees on multiple-choice board-style exams. However, to develop clinically useful multimodal LLM tools, high-quality benchmarks curated by domain experts are essential. To curate released and holdout datasets of 100 chest radiographic studies each and propose an artificial intelligence (AI)-assisted expert labeling procedure to allow radiologists to label studies more efficiently. A total of 13,735 deidentified chest radiographs and their corresponding reports from the MIDRC were used. GPT-4o extracted abnormal findings from the reports, which were then mapped to 12 benchmark labels with a locally hosted LLM (Phi-4-Reasoning). From these studies, 1,000 were sampled on the basis of the AI-suggested benchmark labels for expert review; the sampling algorithm ensured that the selected studies were clinically relevant and captured a range of difficulty levels. Seventeen chest radiologists participated, and they marked "Agree all", "Agree mostly" or "Disagree" to indicate their assessment of the correctness of the LLM suggested labels. Each chest radiograph was evaluated by three experts. Of these, at least two radiologists selected "Agree All" for 381 radiographs. From this set, 200 were selected, prioritizing those with less common or multiple finding labels, and divided into 100 released radiographs and 100 reserved as the holdout dataset. The holdout dataset is used exclusively by RSNA to independently evaluate different models. A benchmark of 200 chest radiographic studies with 12 benchmark labels was created and made publicly available https://imaging.rsna.org, with each chest radiograph verified by three radiologists. In addition, an AI-assisted labeling procedure was developed to help radiologists label at scale, minimize unnecessary omissions, and support a semicollaborative environment.
Learning-Based Radiomic Prediction of Type 2 Diabetes Mellitus Using Image-Derived Phenotypes
Sep 20, 2022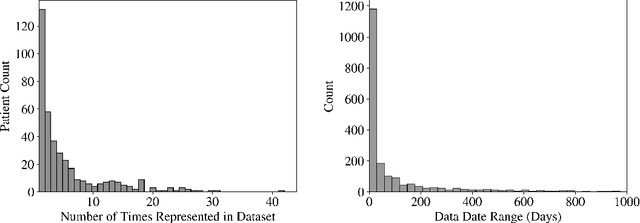
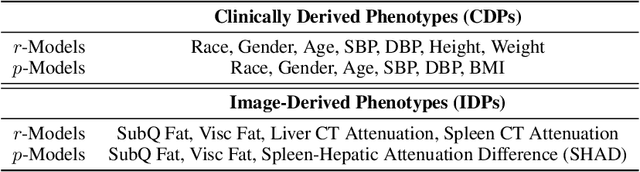
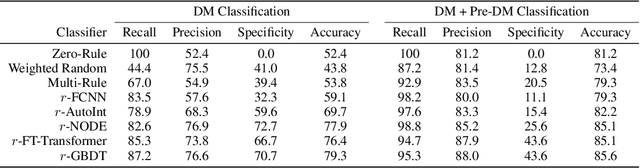
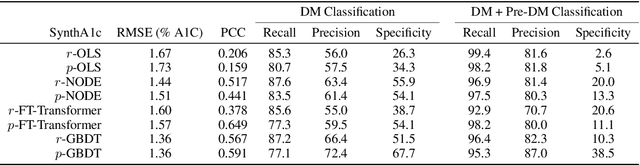
Abstract:Early diagnosis of Type 2 Diabetes Mellitus (T2DM) is crucial to enable timely therapeutic interventions and lifestyle modifications. As medical imaging data become more widely available for many patient populations, we sought to investigate whether image-derived phenotypic data could be leveraged in tabular learning classifier models to predict T2DM incidence without the use of invasive blood lab measurements. We show that both neural network and decision tree models that use image-derived phenotypes can predict patient T2DM status with recall scores as high as 87.6%. We also propose the novel use of these same architectures as 'SynthA1c encoders' that are able to output interpretable values mimicking blood hemoglobin A1C empirical lab measurements. Finally, we demonstrate that T2DM risk prediction model sensitivity to small perturbations in input vector components can be used to predict performance on covariates sampled from previously unseen patient populations.
Deep Learning for Musculoskeletal Image Analysis
Mar 01, 2020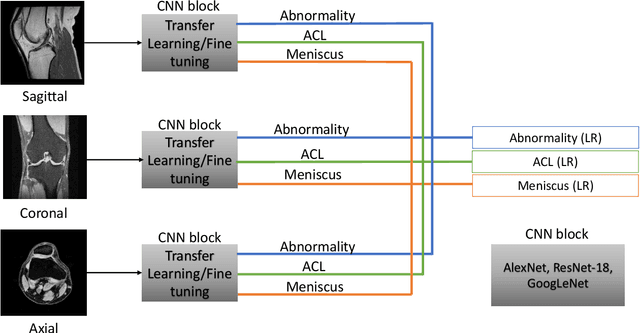
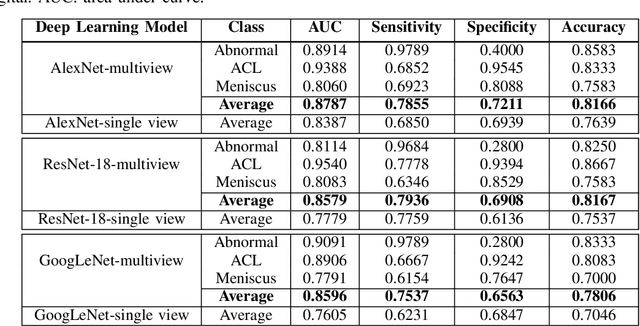
Abstract:The diagnosis, prognosis, and treatment of patients with musculoskeletal (MSK) disorders require radiology imaging (using computed tomography, magnetic resonance imaging(MRI), and ultrasound) and their precise analysis by expert radiologists. Radiology scans can also help assessment of metabolic health, aging, and diabetes. This study presents how machinelearning, specifically deep learning methods, can be used for rapidand accurate image analysis of MRI scans, an unmet clinicalneed in MSK radiology. As a challenging example, we focus on automatic analysis of knee images from MRI scans and study machine learning classification of various abnormalities including meniscus and anterior cruciate ligament tears. Using widely used convolutional neural network (CNN) based architectures, we comparatively evaluated the knee abnormality classification performances of different neural network architectures under limited imaging data regime and compared single and multi-view imaging when classifying the abnormalities. Promising results indicated the potential use of multi-view deep learning based classification of MSK abnormalities in routine clinical assessment.
Weakly Supervised Segmentation by A Deep Geodesic Prior
Aug 18, 2019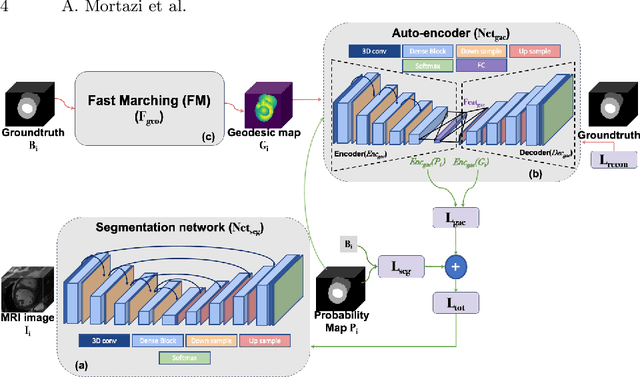
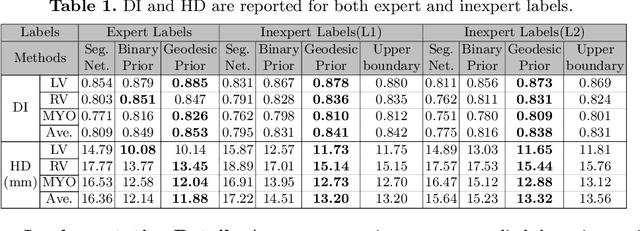
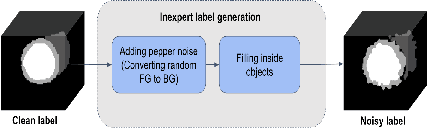
Abstract:The performance of the state-of-the-art image segmentation methods heavily relies on the high-quality annotations, which are not easily affordable, particularly for medical data. To alleviate this limitation, in this study, we propose a weakly supervised image segmentation method based on a deep geodesic prior. We hypothesize that integration of this prior information can reduce the adverse effects of weak labels in segmentation accuracy. Our proposed algorithm is based on a prior information, extracted from an auto-encoder, trained to map objects geodesic maps to their corresponding binary maps. The obtained information is then used as an extra term in the loss function of the segmentor. In order to show efficacy of the proposed strategy, we have experimented segmentation of cardiac substructures with clean and two levels of noisy labels (L1, L2). Our experiments showed that the proposed algorithm boosted the performance of baseline deep learning-based segmentation for both clean and noisy labels by 4.4%, 4.6%(L1), and 6.3%(L2) in dice score, respectively. We also showed that the proposed method was more robust in the presence of high-level noise due to the existence of shape priors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge