Miaomiao Zhang
Design of JiuTian Intelligent Network Simulation Platform
Sep 28, 2023
Abstract:This paper introduced the JiuTian Intelligent Network Simulation Platform, which can provide wireless communication simulation data services for the Open Innovation Platform. The platform contains a series of scalable simulator functionalities, offering open services that enable users to use reinforcement learning algorithms for model training and inference based on simulation environments and data. Additionally, it allows users to address optimization tasks in different scenarios by uploading and updating parameter configurations. The platform and its open services were primarily introduced from the perspectives of background, overall architecture, simulator, business scenarios, and future directions.
SADIR: Shape-Aware Diffusion Models for 3D Image Reconstruction
Sep 06, 2023Abstract:3D image reconstruction from a limited number of 2D images has been a long-standing challenge in computer vision and image analysis. While deep learning-based approaches have achieved impressive performance in this area, existing deep networks often fail to effectively utilize the shape structures of objects presented in images. As a result, the topology of reconstructed objects may not be well preserved, leading to the presence of artifacts such as discontinuities, holes, or mismatched connections between different parts. In this paper, we propose a shape-aware network based on diffusion models for 3D image reconstruction, named SADIR, to address these issues. In contrast to previous methods that primarily rely on spatial correlations of image intensities for 3D reconstruction, our model leverages shape priors learned from the training data to guide the reconstruction process. To achieve this, we develop a joint learning network that simultaneously learns a mean shape under deformation models. Each reconstructed image is then considered as a deformed variant of the mean shape. We validate our model, SADIR, on both brain and cardiac magnetic resonance images (MRIs). Experimental results show that our method outperforms the baselines with lower reconstruction error and better preservation of the shape structure of objects within the images.
NeurEPDiff: Neural Operators to Predict Geodesics in Deformation Spaces
Mar 13, 2023



Abstract:This paper presents NeurEPDiff, a novel network to fast predict the geodesics in deformation spaces generated by a well known Euler-Poincar\'e differential equation (EPDiff). To achieve this, we develop a neural operator that for the first time learns the evolving trajectory of geodesic deformations parameterized in the tangent space of diffeomorphisms(a.k.a velocity fields). In contrast to previous methods that purely fit the training images, our proposed NeurEPDiff learns a nonlinear mapping function between the time-dependent velocity fields. A composition of integral operators and smooth activation functions is formulated in each layer of NeurEPDiff to effectively approximate such mappings. The fact that NeurEPDiff is able to rapidly provide the numerical solution of EPDiff (given any initial condition) results in a significantly reduced computational cost of geodesic shooting of diffeomorphisms in a high-dimensional image space. Additionally, the properties of discretiztion/resolution-invariant of NeurEPDiff make its performance generalizable to multiple image resolutions after being trained offline. We demonstrate the effectiveness of NeurEPDiff in registering two image datasets: 2D synthetic data and 3D brain resonance imaging (MRI). The registration accuracy and computational efficiency are compared with the state-of-the-art diffeomophic registration algorithms with geodesic shooting.
MetaMorph: Learning Metamorphic Image Transformation With Appearance Changes
Mar 08, 2023Abstract:This paper presents a novel predictive model, MetaMorph, for metamorphic registration of images with appearance changes (i.e., caused by brain tumors). In contrast to previous learning-based registration methods that have little or no control over appearance-changes, our model introduces a new regularization that can effectively suppress the negative effects of appearance changing areas. In particular, we develop a piecewise regularization on the tangent space of diffeomorphic transformations (also known as initial velocity fields) via learned segmentation maps of abnormal regions. The geometric transformation and appearance changes are treated as joint tasks that are mutually beneficial. Our model MetaMorph is more robust and accurate when searching for an optimal registration solution under the guidance of segmentation, which in turn improves the segmentation performance by providing appropriately augmented training labels. We validate MetaMorph on real 3D human brain tumor magnetic resonance imaging (MRI) scans. Experimental results show that our model outperforms the state-of-the-art learning-based registration models. The proposed MetaMorph has great potential in various image-guided clinical interventions, e.g., real-time image-guided navigation systems for tumor removal surgery.
Multimodal Deep Learning to Differentiate Tumor Recurrence from Treatment Effect in Human Glioblastoma
Feb 27, 2023Abstract:Differentiating tumor progression (TP) from treatment-related necrosis (TN) is critical for clinical management decisions in glioblastoma (GBM). Dynamic FDG PET (dPET), an advance from traditional static FDG PET, may prove advantageous in clinical staging. dPET includes novel methods of a model-corrected blood input function that accounts for partial volume averaging to compute parametric maps that reveal kinetic information. In a preliminary study, a convolution neural network (CNN) was trained to predict classification accuracy between TP and TN for $35$ brain tumors from $26$ subjects in the PET-MR image space. 3D parametric PET Ki (from dPET), traditional static PET standardized uptake values (SUV), and also the brain tumor MR voxels formed the input for the CNN. The average test accuracy across all leave-one-out cross-validation iterations adjusting for class weights was $0.56$ using only the MR, $0.65$ using only the SUV, and $0.71$ using only the Ki voxels. Combining SUV and MR voxels increased the test accuracy to $0.62$. On the other hand, MR and Ki voxels increased the test accuracy to $0.74$. Thus, dPET features alone or with MR features in deep learning models would enhance prediction accuracy in differentiating TP vs TN in GBM.
Multitask Learning for Improved Late Mechanical Activation Detection of Heart from Cine DENSE MRI
Nov 11, 2022



Abstract:The selection of an optimal pacing site, which is ideally scar-free and late activated, is critical to the response of cardiac resynchronization therapy (CRT). Despite the success of current approaches formulating the detection of such late mechanical activation (LMA) regions as a problem of activation time regression, their accuracy remains unsatisfactory, particularly in cases where myocardial scar exists. To address this issue, this paper introduces a multi-task deep learning framework that simultaneously estimates LMA amount and classify the scar-free LMA regions based on cine displacement encoding with stimulated echoes (DENSE) magnetic resonance imaging (MRI). With a newly introduced auxiliary LMA region classification sub-network, our proposed model shows more robustness to the complex pattern cause by myocardial scar, significantly eliminates their negative effects in LMA detection, and in turn improves the performance of scar classification. To evaluate the effectiveness of our method, we tests our model on real cardiac MR images and compare the predicted LMA with the state-of-the-art approaches. It shows that our approach achieves substantially increased accuracy. In addition, we employ the gradient-weighted class activation mapping (Grad-CAM) to visualize the feature maps learned by all methods. Experimental results suggest that our proposed model better recognizes the LMA region pattern.
Joint Deep Learning for Improved Myocardial Scar Detection from Cardiac MRI
Nov 11, 2022Abstract:Automated identification of myocardial scar from late gadolinium enhancement cardiac magnetic resonance images (LGE-CMR) is limited by image noise and artifacts such as those related to motion and partial volume effect. This paper presents a novel joint deep learning (JDL) framework that improves such tasks by utilizing simultaneously learned myocardium segmentations to eliminate negative effects from non-region-of-interest areas. In contrast to previous approaches treating scar detection and myocardium segmentation as separate or parallel tasks, our proposed method introduces a message passing module where the information of myocardium segmentation is directly passed to guide scar detectors. This newly designed network will efficiently exploit joint information from the two related tasks and use all available sources of myocardium segmentation to benefit scar identification. We demonstrate the effectiveness of JDL on LGE-CMR images for automated left ventricular (LV) scar detection, with great potential to improve risk prediction in patients with both ischemic and non-ischemic heart disease and to improve response rates to cardiac resynchronization therapy (CRT) for heart failure patients. Experimental results show that our proposed approach outperforms multiple state-of-the-art methods, including commonly used two-step segmentation-classification networks, and multitask learning schemes where subtasks are indirectly interacted.
Geo-SIC: Learning Deformable Geometric Shapes in Deep Image Classifiers
Oct 25, 2022Abstract:Deformable shapes provide important and complex geometric features of objects presented in images. However, such information is oftentimes missing or underutilized as implicit knowledge in many image analysis tasks. This paper presents Geo-SIC, the first deep learning model to learn deformable shapes in a deformation space for an improved performance of image classification. We introduce a newly designed framework that (i) simultaneously derives features from both image and latent shape spaces with large intra-class variations; and (ii) gains increased model interpretability by allowing direct access to the underlying geometric features of image data. In particular, we develop a boosted classification network, equipped with an unsupervised learning of geometric shape representations characterized by diffeomorphic transformations within each class. In contrast to previous approaches using pre-extracted shapes, our model provides a more fundamental approach by naturally learning the most relevant shape features jointly with an image classifier. We demonstrate the effectiveness of our method on both simulated 2D images and real 3D brain magnetic resonance (MR) images. Experimental results show that our model substantially improves the image classification accuracy with an additional benefit of increased model interpretability. Our code is publicly available at https://github.com/jw4hv/Geo-SIC
BDG-Net: Boundary Distribution Guided Network for Accurate Polyp Segmentation
Jan 03, 2022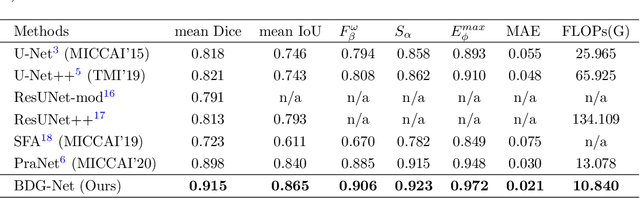
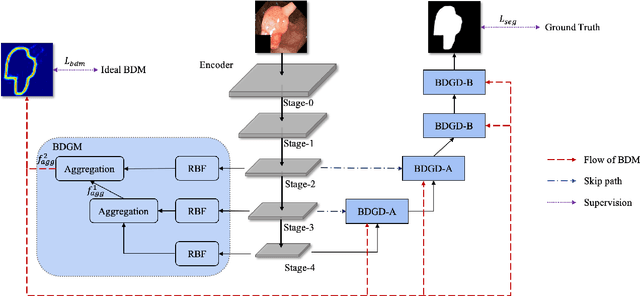
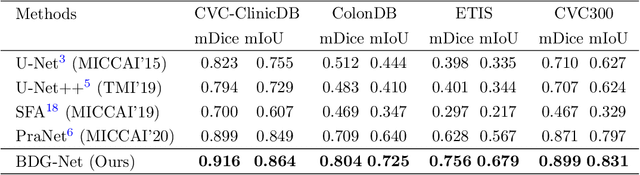
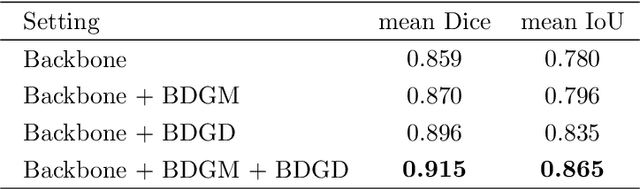
Abstract:Colorectal cancer (CRC) is one of the most common fatal cancer in the world. Polypectomy can effectively interrupt the progression of adenoma to adenocarcinoma, thus reducing the risk of CRC development. Colonoscopy is the primary method to find colonic polyps. However, due to the different sizes of polyps and the unclear boundary between polyps and their surrounding mucosa, it is challenging to segment polyps accurately. To address this problem, we design a Boundary Distribution Guided Network (BDG-Net) for accurate polyp segmentation. Specifically, under the supervision of the ideal Boundary Distribution Map (BDM), we use Boundary Distribution Generate Module (BDGM) to aggregate high-level features and generate BDM. Then, BDM is sent to the Boundary Distribution Guided Decoder (BDGD) as complementary spatial information to guide the polyp segmentation. Moreover, a multi-scale feature interaction strategy is adopted in BDGD to improve the segmentation accuracy of polyps with different sizes. Extensive quantitative and qualitative evaluations demonstrate the effectiveness of our model, which outperforms state-of-the-art models remarkably on five public polyp datasets while maintaining low computational complexity.
Hybrid Atlas Building with Deep Registration Priors
Dec 13, 2021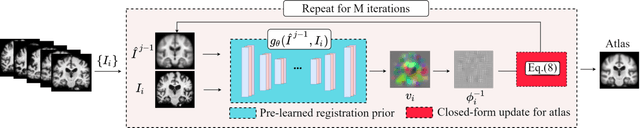
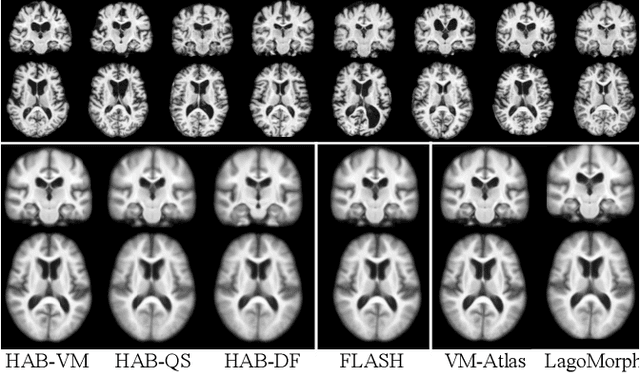
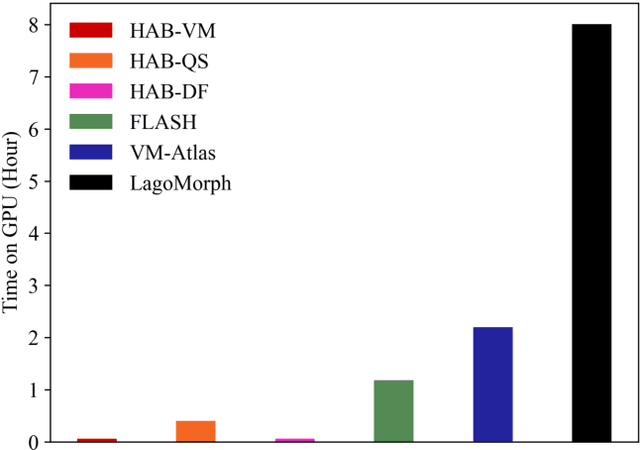
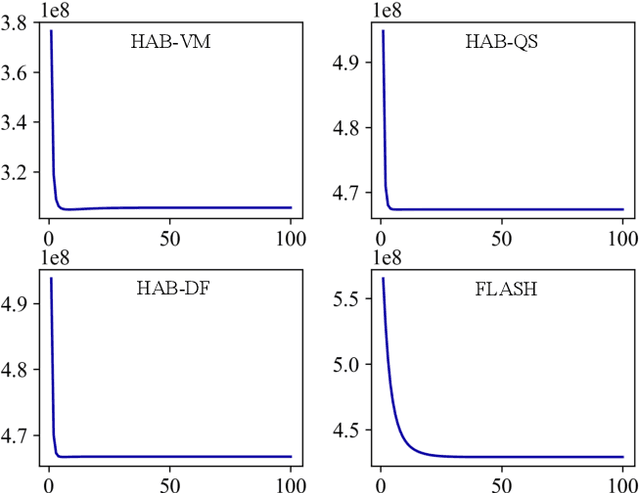
Abstract:Registration-based atlas building often poses computational challenges in high-dimensional image spaces. In this paper, we introduce a novel hybrid atlas building algorithm that fast estimates atlas from large-scale image datasets with much reduced computational cost. In contrast to previous approaches that iteratively perform registration tasks between an estimated atlas and individual images, we propose to use learned priors of registration from pre-trained neural networks. This newly developed hybrid framework features several advantages of (i) providing an efficient way of atlas building without losing the quality of results, and (ii) offering flexibility in utilizing a wide variety of deep learning based registration methods. We demonstrate the effectiveness of this proposed model on 3D brain magnetic resonance imaging (MRI) scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge