Zihuan Qiu
FireRescue: A UAV-Based Dataset and Enhanced YOLO Model for Object Detection in Fire Rescue Scenes
Dec 31, 2025Abstract:Object detection in fire rescue scenarios is importance for command and decision-making in firefighting operations. However, existing research still suffers from two main limitations. First, current work predominantly focuses on environments such as mountainous or forest areas, while paying insufficient attention to urban rescue scenes, which are more frequent and structurally complex. Second, existing detection systems include a limited number of classes, such as flames and smoke, and lack a comprehensive system covering key targets crucial for command decisions, such as fire trucks and firefighters. To address the above issues, this paper first constructs a new dataset named "FireRescue" for rescue command, which covers multiple rescue scenarios, including urban, mountainous, forest, and water areas, and contains eight key categories such as fire trucks and firefighters, with a total of 15,980 images and 32,000 bounding boxes. Secondly, to tackle the problems of inter-class confusion and missed detection of small targets caused by chaotic scenes, diverse targets, and long-distance shooting, this paper proposes an improved model named FRS-YOLO. On the one hand, the model introduces a plug-and-play multidi-mensional collaborative enhancement attention module, which enhances the discriminative representation of easily confused categories (e.g., fire trucks vs. ordinary trucks) through cross-dimensional feature interaction. On the other hand, it integrates a dynamic feature sampler to strengthen high-response foreground features, thereby mitigating the effects of smoke occlusion and background interference. Experimental results demonstrate that object detection in fire rescue scenarios is highly challenging, and the proposed method effectively improves the detection performance of YOLO series models in this context.
Closing the Oracle Gap: Increment Vector Transformation for Class Incremental Learning
Sep 26, 2025Abstract:Class Incremental Learning (CIL) aims to sequentially acquire knowledge of new classes without forgetting previously learned ones. Despite recent progress, current CIL methods still exhibit significant performance gaps compared to their oracle counterparts-models trained with full access to historical data. Inspired by recent insights on Linear Mode Connectivity (LMC), we revisit the geometric properties of oracle solutions in CIL and uncover a fundamental observation: these oracle solutions typically maintain low-loss linear connections to the optimum of previous tasks. Motivated by this finding, we propose Increment Vector Transformation (IVT), a novel plug-and-play framework designed to mitigate catastrophic forgetting during training. Rather than directly following CIL updates, IVT periodically teleports the model parameters to transformed solutions that preserve linear connectivity to previous task optimum. By maintaining low-loss along these connecting paths, IVT effectively ensures stable performance on previously learned tasks. The transformation is efficiently approximated using diagonal Fisher Information Matrices, making IVT suitable for both exemplar-free and exemplar-based scenarios, and compatible with various initialization strategies. Extensive experiments on CIFAR-100, FGVCAircraft, ImageNet-Subset, and ImageNet-Full demonstrate that IVT consistently enhances the performance of strong CIL baselines. Specifically, on CIFAR-100, IVT improves the last accuracy of the PASS baseline by +5.12% and reduces forgetting by 2.54%. For the CLIP-pre-trained SLCA baseline on FGVCAircraft, IVT yields gains of +14.93% in average accuracy and +21.95% in last accuracy. The code will be released.
MINGLE: Mixtures of Null-Space Gated Low-Rank Experts for Test-Time Continual Model Merging
May 17, 2025Abstract:Continual model merging integrates independently fine-tuned models sequentially without access to original training data, providing a scalable and efficient solution to continual learning. However, current methods still face critical challenges, notably parameter interference among tasks and limited adaptability to evolving test distributions. The former causes catastrophic forgetting of integrated tasks, while the latter hinders effective adaptation to new tasks. To address these, we propose MINGLE, a novel framework for test-time continual model merging, which leverages test-time adaptation using a small set of unlabeled test samples from the current task to dynamically guide the merging process. MINGLE employs a mixture-of-experts architecture composed of parameter-efficient, low-rank experts, enabling efficient adaptation and improving robustness to distribution shifts. To mitigate catastrophic forgetting, we propose Null-Space Constrained Gating, which restricts gating updates to subspaces orthogonal to prior task representations. This suppresses activations on old task inputs and preserves model behavior on past tasks. To further balance stability and adaptability, we design an Adaptive Relaxation Strategy, which dynamically adjusts the constraint strength based on interference signals captured during test-time adaptation. Extensive experiments on standard continual merging benchmarks demonstrate that MINGLE achieves robust generalization, reduces forgetting significantly, and consistently surpasses previous state-of-the-art methods by 7-9\% on average across diverse task orders.
Distribution-Level Memory Recall for Continual Learning: Preserving Knowledge and Avoiding Confusion
Aug 04, 2024



Abstract:Continual Learning (CL) aims to enable Deep Neural Networks (DNNs) to learn new data without forgetting previously learned knowledge. The key to achieving this goal is to avoid confusion at the feature level, i.e., avoiding confusion within old tasks and between new and old tasks. Previous prototype-based CL methods generate pseudo features for old knowledge replay by adding Gaussian noise to the centroids of old classes. However, the distribution in the feature space exhibits anisotropy during the incremental process, which prevents the pseudo features from faithfully reproducing the distribution of old knowledge in the feature space, leading to confusion in classification boundaries within old tasks. To address this issue, we propose the Distribution-Level Memory Recall (DMR) method, which uses a Gaussian mixture model to precisely fit the feature distribution of old knowledge at the distribution level and generate pseudo features in the next stage. Furthermore, resistance to confusion at the distribution level is also crucial for multimodal learning, as the problem of multimodal imbalance results in significant differences in feature responses between different modalities, exacerbating confusion within old tasks in prototype-based CL methods. Therefore, we mitigate the multi-modal imbalance problem by using the Inter-modal Guidance and Intra-modal Mining (IGIM) method to guide weaker modalities with prior information from dominant modalities and further explore useful information within modalities. For the second key, We propose the Confusion Index to quantitatively describe a model's ability to distinguish between new and old tasks, and we use the Incremental Mixup Feature Enhancement (IMFE) method to enhance pseudo features with new sample features, alleviating classification confusion between new and old knowledge.
BDG-Net: Boundary Distribution Guided Network for Accurate Polyp Segmentation
Jan 03, 2022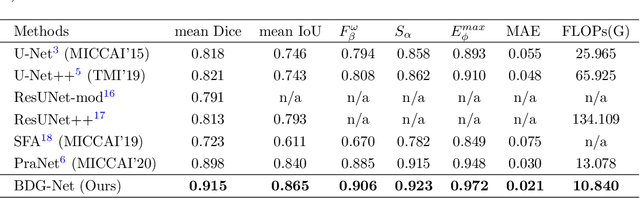
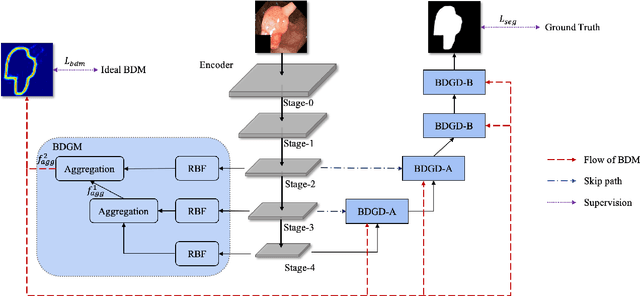
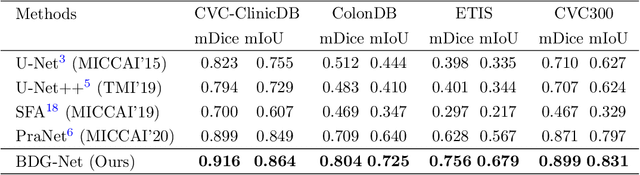
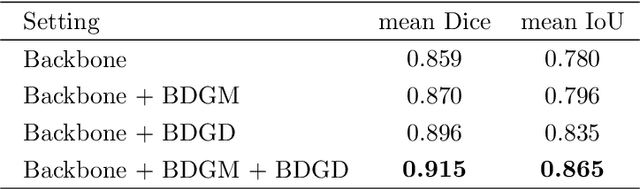
Abstract:Colorectal cancer (CRC) is one of the most common fatal cancer in the world. Polypectomy can effectively interrupt the progression of adenoma to adenocarcinoma, thus reducing the risk of CRC development. Colonoscopy is the primary method to find colonic polyps. However, due to the different sizes of polyps and the unclear boundary between polyps and their surrounding mucosa, it is challenging to segment polyps accurately. To address this problem, we design a Boundary Distribution Guided Network (BDG-Net) for accurate polyp segmentation. Specifically, under the supervision of the ideal Boundary Distribution Map (BDM), we use Boundary Distribution Generate Module (BDGM) to aggregate high-level features and generate BDM. Then, BDM is sent to the Boundary Distribution Guided Decoder (BDGD) as complementary spatial information to guide the polyp segmentation. Moreover, a multi-scale feature interaction strategy is adopted in BDGD to improve the segmentation accuracy of polyps with different sizes. Extensive quantitative and qualitative evaluations demonstrate the effectiveness of our model, which outperforms state-of-the-art models remarkably on five public polyp datasets while maintaining low computational complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge