Marleen de Bruijne
for the ALFA study
A Unified Framework for Joint Detection of Lacunes and Enlarged Perivascular Spaces
Mar 05, 2026Abstract:Cerebral small vessel disease (CSVD) markers, specifically enlarged perivascular spaces (EPVS) and lacunae, present a unique challenge in medical image analysis due to their radiological mimicry. Standard segmentation networks struggle with feature interference and extreme class imbalance when handling these divergent targets simultaneously. To address these issues, we propose a morphology-decoupled framework where Zero-Initialized Gated Cross-Task Attention exploits dense EPVS context to guide sparse lacune detection. Furthermore, biological and topological consistency are enforced via a mixed-supervision strategy integrating Mutual Exclusion and Centerline Dice losses. Finally, we introduce an Anatomically-Informed Inference Calibration mechanism to dynamically suppress false positives based on tissue semantics. Extensive 5-folds cross-validation on the VALDO 2021 dataset (N=40) demonstrates state-of-the-art performance, notably surpassing task winners in lacunae detection precision (71.1%, p=0.01) and F1-score (62.6%, p=0.03). Furthermore, evaluation on the external EPAD cohort (N=1762) confirms the model's robustness for large-scale population studies. Code will be released upon acceptance.
rNCA: Self-Repairing Segmentation Masks
Dec 15, 2025Abstract:Accurately predicting topologically correct masks remains a difficult task for general segmentation models, which often produce fragmented or disconnected outputs. Fixing these artifacts typically requires hand-crafted refinement rules or architectures specialized to a particular task. Here, we show that Neural Cellular Automata (NCA) can be directly re-purposed as an effective refinement mechanism, using local, iterative updates guided by image context to repair segmentation masks. By training on imperfect masks and ground truths, the automaton learns the structural properties of the target shape while relying solely on local information. When applied to coarse, globally predicted masks, the learned dynamics progressively reconnect broken regions, prune loose fragments and converge towards stable, topologically consistent results. We show how refinement NCA (rNCA) can be easily applied to repair common topological errors produced by different base segmentation models and tasks: for fragmented retinal vessels, it yields 2-3% gains in Dice/clDice and improves Betti errors, reducing $β_0$ errors by 60% and $β_1$ by 20%; for myocardium, it repairs 61.5% of broken cases in a zero-shot setting while lowering ASSD and HD by 19% and 16%, respectively. This showcases NCA as effective and broadly applicable refiners.
Leveraging point annotations in segmentation learning with boundary loss
Nov 06, 2023Abstract:This paper investigates the combination of intensity-based distance maps with boundary loss for point-supervised semantic segmentation. By design the boundary loss imposes a stronger penalty on the false positives the farther away from the object they occur. Hence it is intuitively inappropriate for weak supervision, where the ground truth label may be much smaller than the actual object and a certain amount of false positives (w.r.t. the weak ground truth) is actually desirable. Using intensity-aware distances instead may alleviate this drawback, allowing for a certain amount of false positives without a significant increase to the training loss. The motivation for applying the boundary loss directly under weak supervision lies in its great success for fully supervised segmentation tasks, but also in not requiring extra priors or outside information that is usually required -- in some form -- with existing weakly supervised methods in the literature. This formulation also remains potentially more attractive than existing CRF-based regularizers, due to its simplicity and computational efficiency. We perform experiments on two multi-class datasets; ACDC (heart segmentation) and POEM (whole-body abdominal organ segmentation). Preliminary results are encouraging and show that this supervision strategy has great potential. On ACDC it outperforms the CRF-loss based approach, and on POEM data it performs on par with it. The code for all our experiments is openly available.
Source Identification: A Self-Supervision Task for Dense Prediction
Jul 05, 2023



Abstract:The paradigm of self-supervision focuses on representation learning from raw data without the need of labor-consuming annotations, which is the main bottleneck of current data-driven methods. Self-supervision tasks are often used to pre-train a neural network with a large amount of unlabeled data and extract generic features of the dataset. The learned model is likely to contain useful information which can be transferred to the downstream main task and improve performance compared to random parameter initialization. In this paper, we propose a new self-supervision task called source identification (SI), which is inspired by the classic blind source separation problem. Synthetic images are generated by fusing multiple source images and the network's task is to reconstruct the original images, given the fused images. A proper understanding of the image content is required to successfully solve the task. We validate our method on two medical image segmentation tasks: brain tumor segmentation and white matter hyperintensities segmentation. The results show that the proposed SI task outperforms traditional self-supervision tasks for dense predictions including inpainting, pixel shuffling, intensity shift, and super-resolution. Among variations of the SI task fusing images of different types, fusing images from different patients performs best.
On the dice loss gradient and the ways to mimic it
Apr 09, 2023



Abstract:In the past few years, in the context of fully-supervised semantic segmentation, several losses -- such as cross-entropy and dice -- have emerged as de facto standards to supervise neural networks. The Dice loss is an interesting case, as it comes from the relaxation of the popular Dice coefficient; one of the main evaluation metric in medical imaging applications. In this paper, we first study theoretically the gradient of the dice loss, showing that concretely it is a weighted negative of the ground truth, with a very small dynamic range. This enables us, in the second part of this paper, to mimic the supervision of the dice loss, through a simple element-wise multiplication of the network output with a negative of the ground truth. This rather surprising result sheds light on the practical supervision performed by the dice loss during gradient descent. This can help the practitioner to understand and interpret results while guiding researchers when designing new losses.
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Label Refinement Network from Synthetic Error Augmentation for Medical Image Segmentation
Sep 14, 2022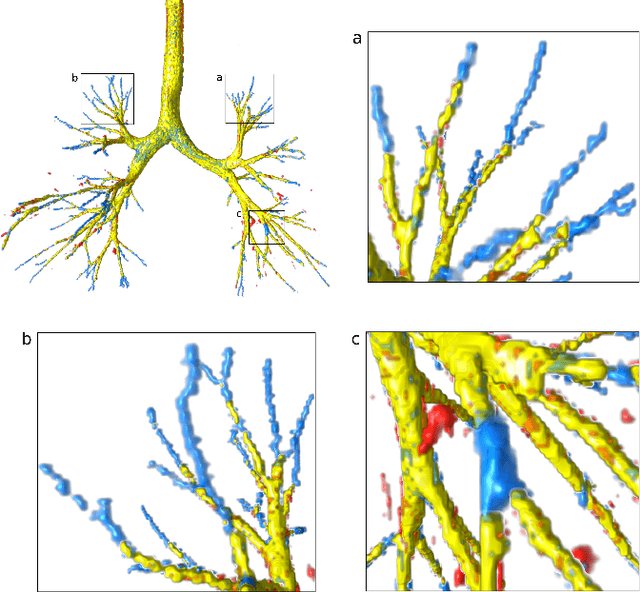
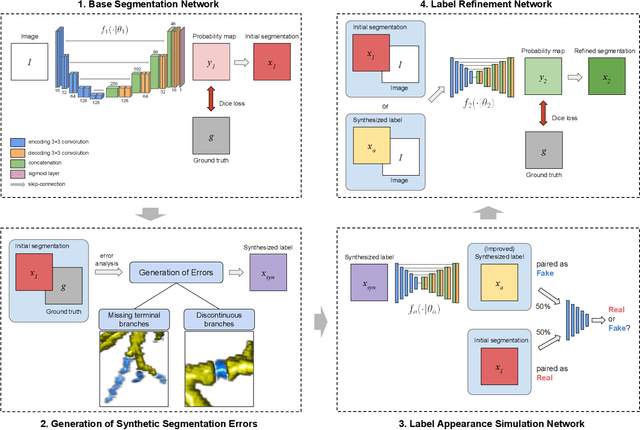
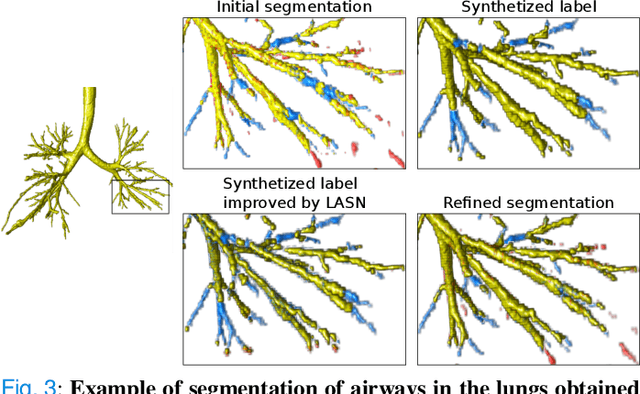
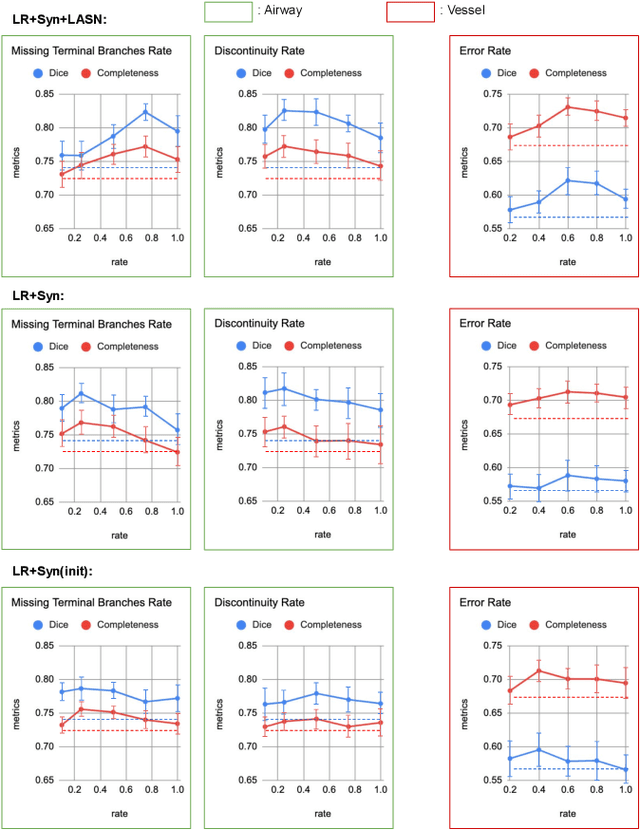
Abstract:Deep convolutional neural networks for image segmentation do not learn the label structure explicitly and may produce segmentations with an incorrect structure, e.g., with disconnected cylindrical structures in the segmentation of tree-like structures such as airways or blood vessels. In this paper, we propose a novel label refinement method to correct such errors from an initial segmentation, implicitly incorporating information about label structure. This method features two novel parts: 1) a model that generates synthetic structural errors, and 2) a label appearance simulation network that produces synthetic segmentations (with errors) that are similar in appearance to the real initial segmentations. Using these synthetic segmentations and the original images, the label refinement network is trained to correct errors and improve the initial segmentations. The proposed method is validated on two segmentation tasks: airway segmentation from chest computed tomography (CT) scans and brain vessel segmentation from 3D CT angiography (CTA) images of the brain. In both applications, our method significantly outperformed a standard 3D U-Net and other previous refinement approaches. Improvements are even larger when additional unlabeled data is used for model training. In an ablation study, we demonstrate the value of the different components of the proposed method.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022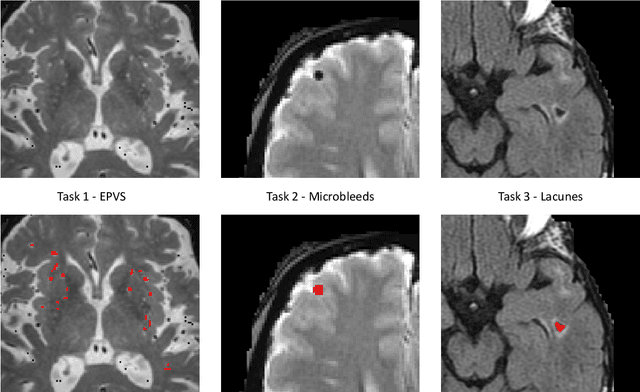
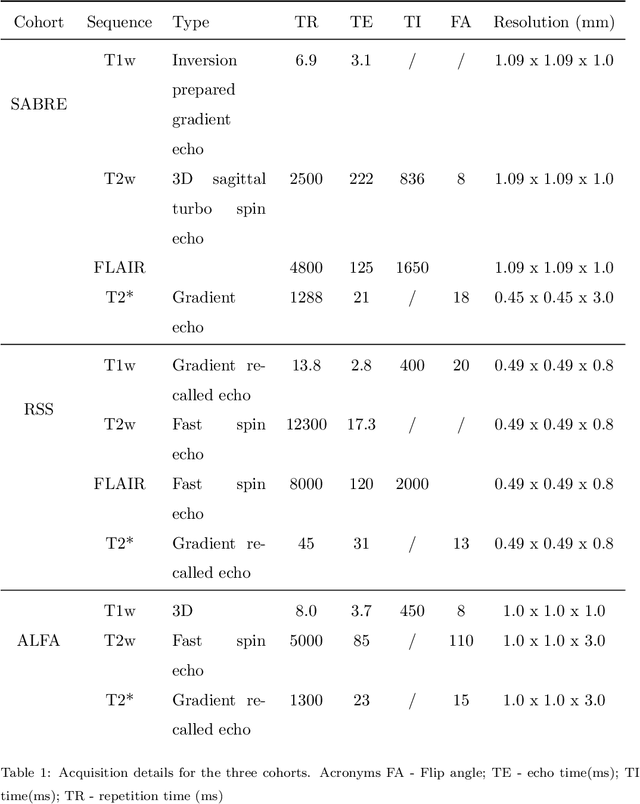
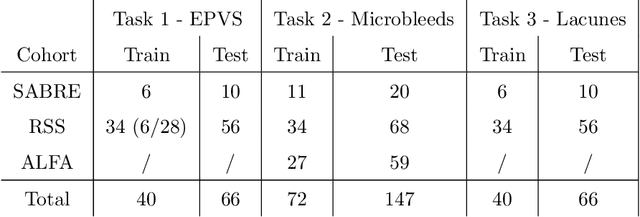
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
A Quantitative Comparison of Epistemic Uncertainty Maps Applied to Multi-Class Segmentation
Sep 22, 2021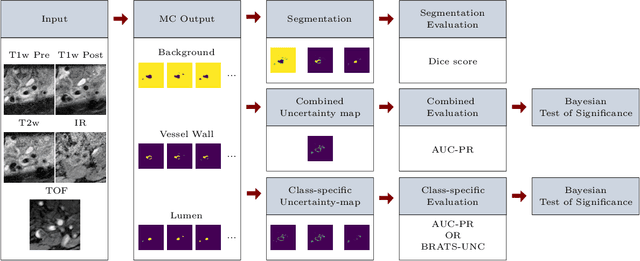
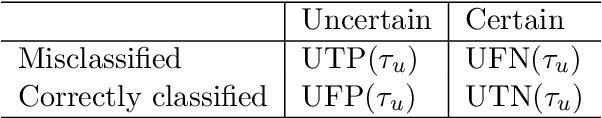
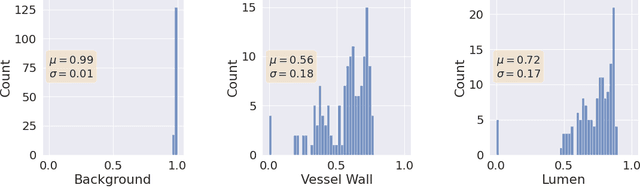
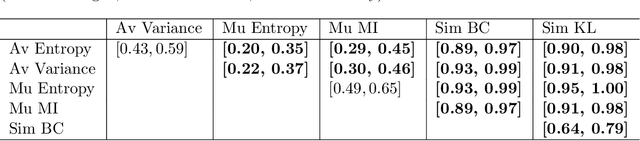
Abstract:Uncertainty assessment has gained rapid interest in medical image analysis. A popular technique to compute epistemic uncertainty is the Monte-Carlo (MC) dropout technique. From a network with MC dropout and a single input, multiple outputs can be sampled. Various methods can be used to obtain epistemic uncertainty maps from those multiple outputs. In the case of multi-class segmentation, the number of methods is even larger as epistemic uncertainty can be computed voxelwise per class or voxelwise per image. This paper highlights a systematic approach to define and quantitatively compare those methods in two different contexts: class-specific epistemic uncertainty maps (one value per image, voxel and class) and combined epistemic uncertainty maps (one value per image and voxel). We applied this quantitative analysis to a multi-class segmentation of the carotid artery lumen and vessel wall, on a multi-center, multi-scanner, multi-sequence dataset of (MR) images. We validated our analysis over 144 sets of hyperparameters of a model. Our main analysis considers the relationship between the order of the voxels sorted according to their epistemic uncertainty values and the misclassification of the prediction. Under this consideration, the comparison of combined uncertainty maps reveals that the multi-class entropy and the multi-class mutual information statistically out-perform the other combined uncertainty maps under study. In a class-specific scenario, the one-versus-all entropy statistically out-performs the class-wise entropy, the class-wise variance and the one versus all mutual information. The class-wise entropy statistically out-performs the other class-specific uncertainty maps in terms of calibration. We made a python package available to reproduce our analysis on different data and tasks.
Deep Learning methods for automatic evaluation of delayed enhancement-MRI. The results of the EMIDEC challenge
Aug 10, 2021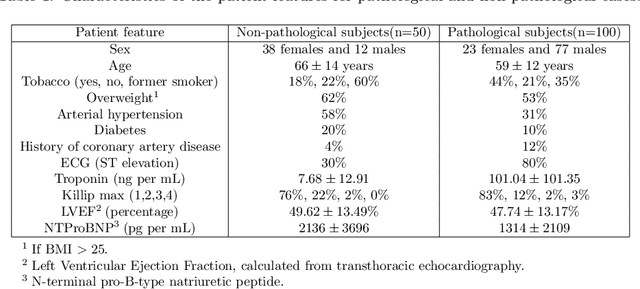
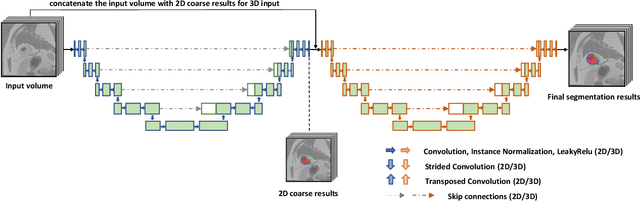
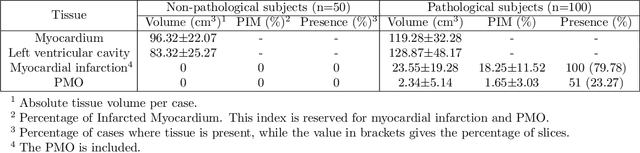
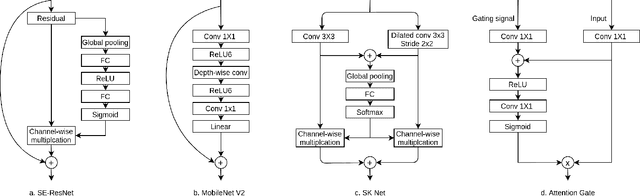
Abstract:A key factor for assessing the state of the heart after myocardial infarction (MI) is to measure whether the myocardium segment is viable after reperfusion or revascularization therapy. Delayed enhancement-MRI or DE-MRI, which is performed several minutes after injection of the contrast agent, provides high contrast between viable and nonviable myocardium and is therefore a method of choice to evaluate the extent of MI. To automatically assess myocardial status, the results of the EMIDEC challenge that focused on this task are presented in this paper. The challenge's main objectives were twofold. First, to evaluate if deep learning methods can distinguish between normal and pathological cases. Second, to automatically calculate the extent of myocardial infarction. The publicly available database consists of 150 exams divided into 50 cases with normal MRI after injection of a contrast agent and 100 cases with myocardial infarction (and then with a hyperenhanced area on DE-MRI), whatever their inclusion in the cardiac emergency department. Along with MRI, clinical characteristics are also provided. The obtained results issued from several works show that the automatic classification of an exam is a reachable task (the best method providing an accuracy of 0.92), and the automatic segmentation of the myocardium is possible. However, the segmentation of the diseased area needs to be improved, mainly due to the small size of these areas and the lack of contrast with the surrounding structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge