Kai Ma
School of Electrical Engineering, Yanshan University, Qinhuangdao, China
Multi-Task Neural Networks with Spatial Activation for Retinal Vessel Segmentation and Artery/Vein Classification
Jul 18, 2020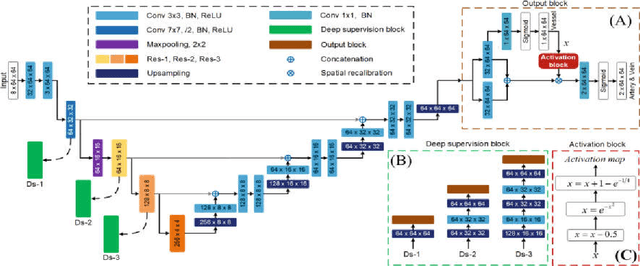
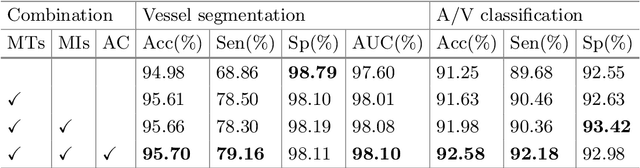
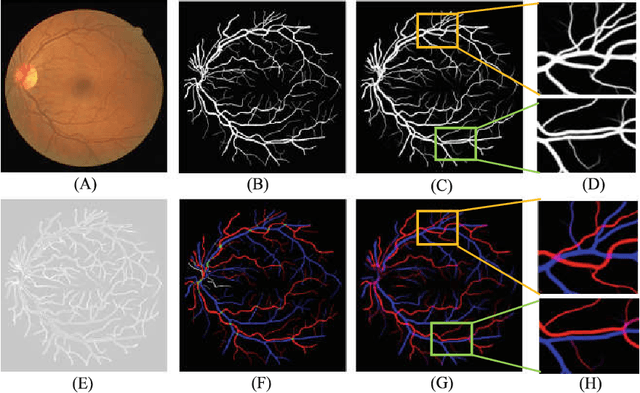
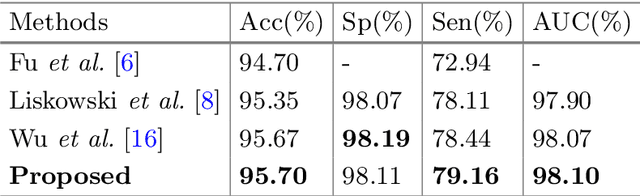
Abstract:Retinal artery/vein (A/V) classification plays a critical role in the clinical biomarker study of how various systemic and cardiovascular diseases affect the retinal vessels. Conventional methods of automated A/V classification are generally complicated and heavily depend on the accurate vessel segmentation. In this paper, we propose a multi-task deep neural network with spatial activation mechanism that is able to segment full retinal vessel, artery and vein simultaneously, without the pre-requirement of vessel segmentation. The input module of the network integrates the domain knowledge of widely used retinal preprocessing and vessel enhancement techniques. We specially customize the output block of the network with a spatial activation mechanism, which takes advantage of a relatively easier task of vessel segmentation and exploits it to boost the performance of A/V classification. In addition, deep supervision is introduced to the network to assist the low level layers to extract more semantic information. The proposed network achieves pixel-wise accuracy of 95.70% for vessel segmentation, and A/V classification accuracy of 94.50%, which is the state-of-the-art performance for both tasks on the AV-DRIVE dataset. Furthermore, we have also tested the model performance on INSPIRE-AVR dataset, which achieves a skeletal A/V classification accuracy of 91.6%.
Superpixel-Guided Label Softening for Medical Image Segmentation
Jul 17, 2020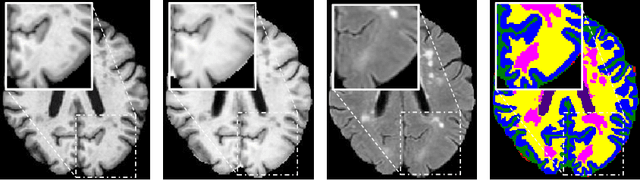
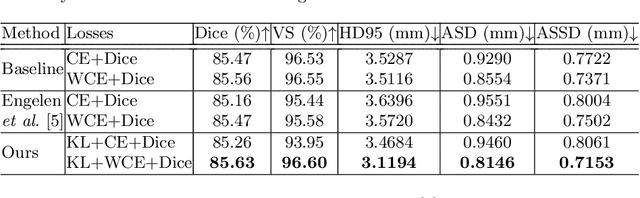
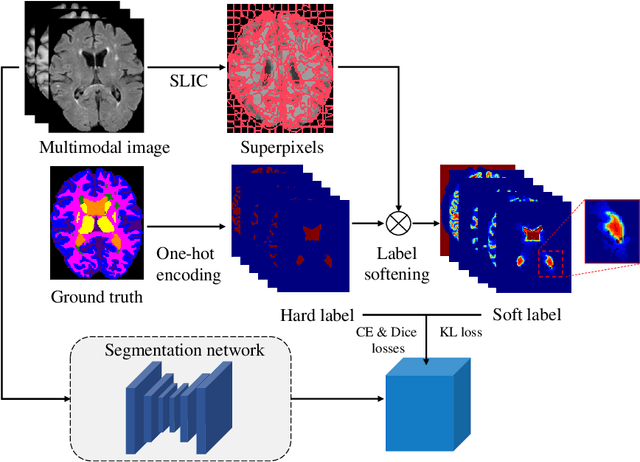
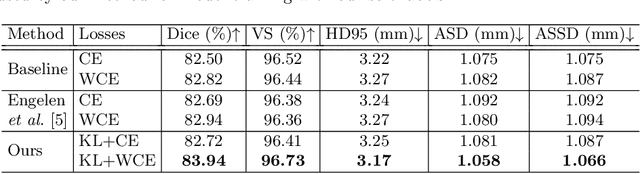
Abstract:Segmentation of objects of interest is one of the central tasks in medical image analysis, which is indispensable for quantitative analysis. When developing machine-learning based methods for automated segmentation, manual annotations are usually used as the ground truth toward which the models learn to mimic. While the bulky parts of the segmentation targets are relatively easy to label, the peripheral areas are often difficult to handle due to ambiguous boundaries and the partial volume effect, etc., and are likely to be labeled with uncertainty. This uncertainty in labeling may, in turn, result in unsatisfactory performance of the trained models. In this paper, we propose superpixel-based label softening to tackle the above issue. Generated by unsupervised over-segmentation, each superpixel is expected to represent a locally homogeneous area. If a superpixel intersects with the annotation boundary, we consider a high probability of uncertain labeling within this area. Driven by this intuition, we soften labels in this area based on signed distances to the annotation boundary and assign probability values within [0, 1] to them, in comparison with the original "hard", binary labels of either 0 or 1. The softened labels are then used to train the segmentation models together with the hard labels. Experimental results on a brain MRI dataset and an optical coherence tomography dataset demonstrate that this conceptually simple and implementation-wise easy method achieves overall superior segmentation performances to baseline and comparison methods for both 3D and 2D medical images.
Revisiting Rubik's Cube: Self-supervised Learning with Volume-wise Transformation for 3D Medical Image Segmentation
Jul 17, 2020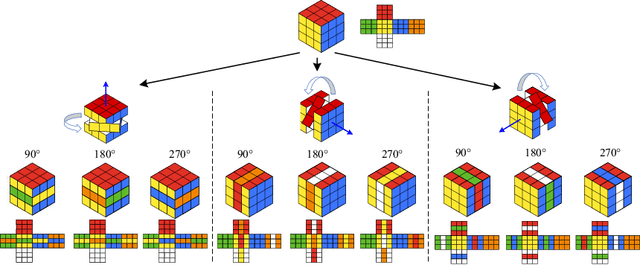
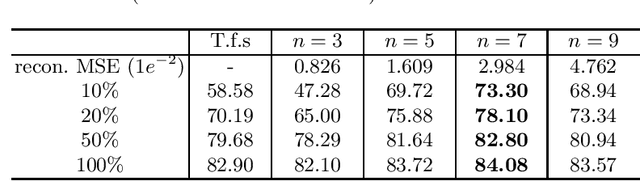
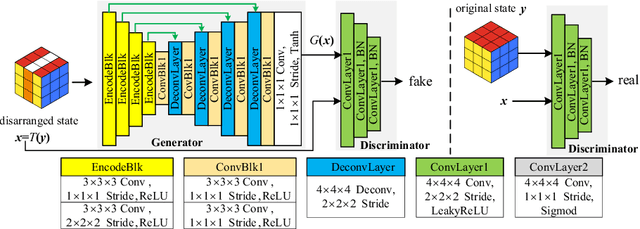
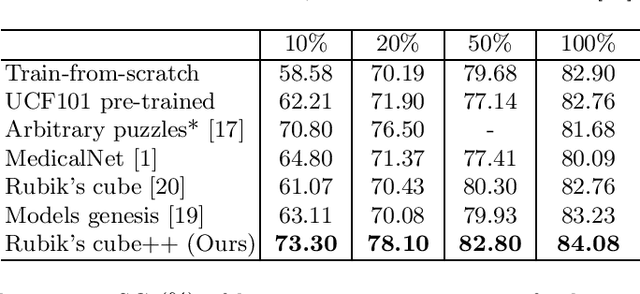
Abstract:Deep learning highly relies on the quantity of annotated data. However, the annotations for 3D volumetric medical data require experienced physicians to spend hours or even days for investigation. Self-supervised learning is a potential solution to get rid of the strong requirement of training data by deeply exploiting raw data information. In this paper, we propose a novel self-supervised learning framework for volumetric medical images. Specifically, we propose a context restoration task, i.e., Rubik's cube++, to pre-train 3D neural networks. Different from the existing context-restoration-based approaches, we adopt a volume-wise transformation for context permutation, which encourages network to better exploit the inherent 3D anatomical information of organs. Compared to the strategy of training from scratch, fine-tuning from the Rubik's cube++ pre-trained weight can achieve better performance in various tasks such as pancreas segmentation and brain tissue segmentation. The experimental results show that our self-supervised learning method can significantly improve the accuracy of 3D deep learning networks on volumetric medical datasets without the use of extra data.
Learning and Exploiting Interclass Visual Correlations for Medical Image Classification
Jul 13, 2020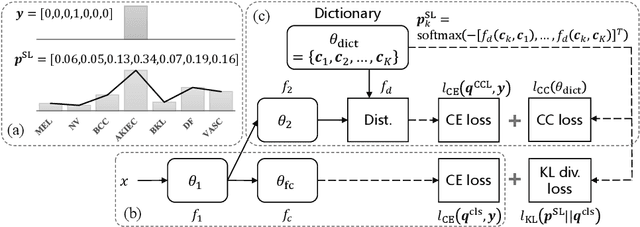
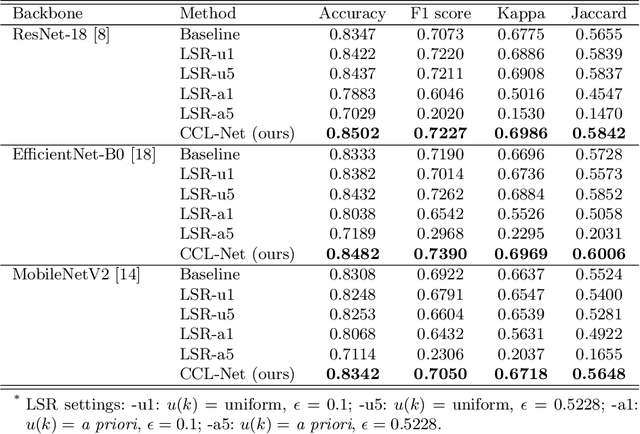
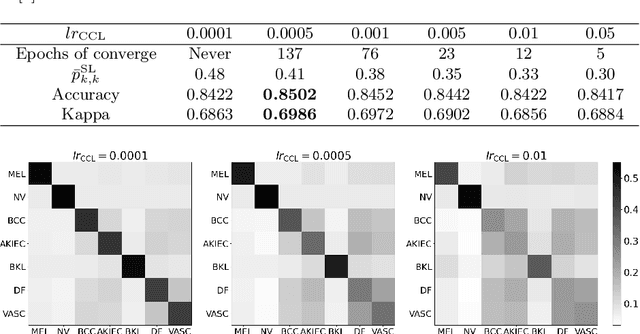
Abstract:Deep neural network-based medical image classifications often use "hard" labels for training, where the probability of the correct category is 1 and those of others are 0. However, these hard targets can drive the networks over-confident about their predictions and prone to overfit the training data, affecting model generalization and adaption. Studies have shown that label smoothing and softening can improve classification performance. Nevertheless, existing approaches are either non-data-driven or limited in applicability. In this paper, we present the Class-Correlation Learning Network (CCL-Net) to learn interclass visual correlations from given training data, and produce soft labels to help with classification tasks. Instead of letting the network directly learn the desired correlations, we propose to learn them implicitly via distance metric learning of class-specific embeddings with a lightweight plugin CCL block. An intuitive loss based on a geometrical explanation of correlation is designed for bolstering learning of the interclass correlations. We further present end-to-end training of the proposed CCL block as a plugin head together with the classification backbone while generating soft labels on the fly. Our experimental results on the International Skin Imaging Collaboration 2018 dataset demonstrate effective learning of the interclass correlations from training data, as well as consistent improvements in performance upon several widely used modern network structures with the CCL block.
Cross-denoising Network against Corrupted Labels in Medical Image Segmentation with Domain Shift
Jun 19, 2020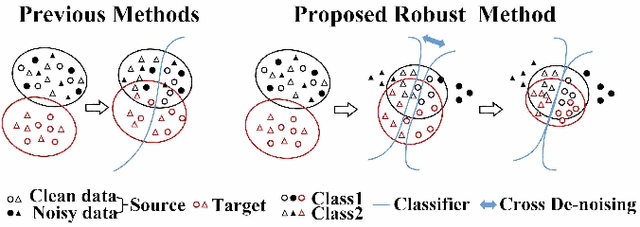
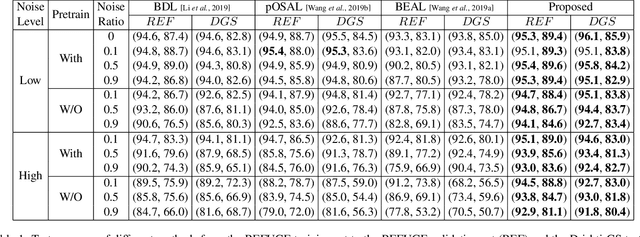
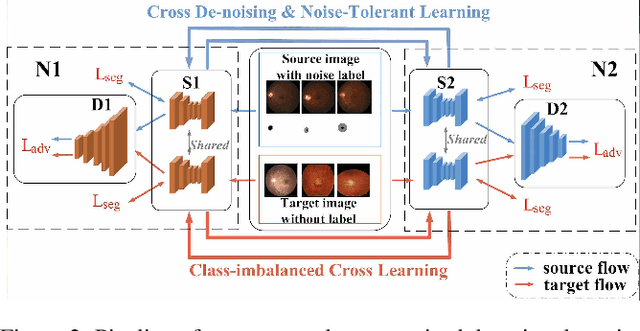
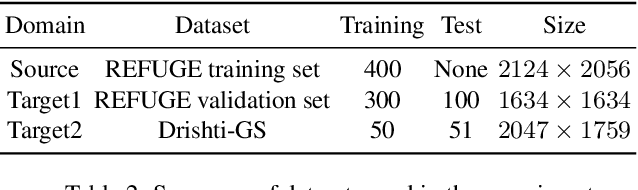
Abstract:Deep convolutional neural networks (DCNNs) have contributed many breakthroughs in segmentation tasks, especially in the field of medical imaging. However, \textit{domain shift} and \textit{corrupted annotations}, which are two common problems in medical imaging, dramatically degrade the performance of DCNNs in practice. In this paper, we propose a novel robust cross-denoising framework using two peer networks to address domain shift and corrupted label problems with a peer-review strategy. Specifically, each network performs as a mentor, mutually supervised to learn from reliable samples selected by the peer network to combat with corrupted labels. In addition, a noise-tolerant loss is proposed to encourage the network to capture the key location and filter the discrepancy under various noise-contaminant labels. To further reduce the accumulated error, we introduce a class-imbalanced cross learning using most confident predictions at the class-level. Experimental results on REFUGE and Drishti-GS datasets for optic disc (OD) and optic cup (OC) segmentation demonstrate the superior performance of our proposed approach to the state-of-the-art methods.
Generative Adversarial Networks for Video-to-Video Domain Adaptation
Apr 17, 2020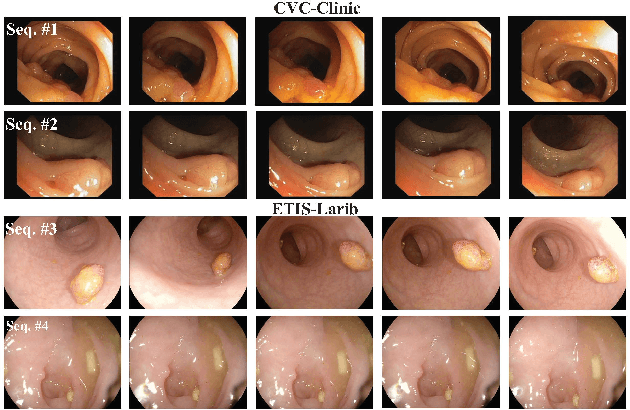
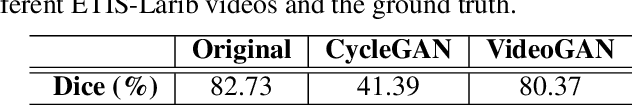
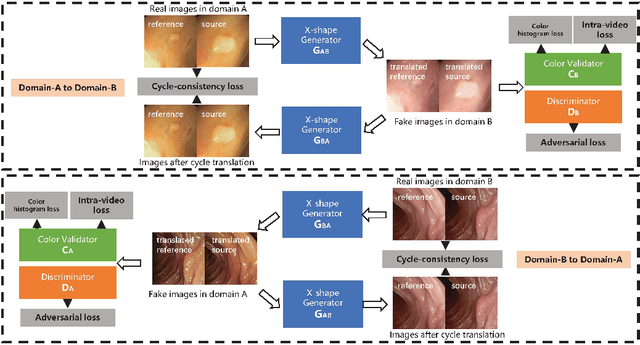
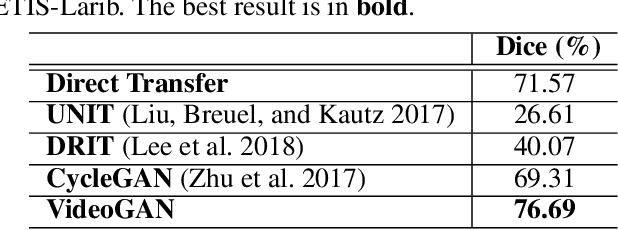
Abstract:Endoscopic videos from multicentres often have different imaging conditions, e.g., color and illumination, which make the models trained on one domain usually fail to generalize well to another. Domain adaptation is one of the potential solutions to address the problem. However, few of existing works focused on the translation of video-based data. In this work, we propose a novel generative adversarial network (GAN), namely VideoGAN, to transfer the video-based data across different domains. As the frames of a video may have similar content and imaging conditions, the proposed VideoGAN has an X-shape generator to preserve the intra-video consistency during translation. Furthermore, a loss function, namely color histogram loss, is proposed to tune the color distribution of each translated frame. Two colonoscopic datasets from different centres, i.e., CVC-Clinic and ETIS-Larib, are adopted to evaluate the performance of domain adaptation of our VideoGAN. Experimental results demonstrate that the adapted colonoscopic video generated by our VideoGAN can significantly boost the segmentation accuracy, i.e., an improvement of 5%, of colorectal polyps on multicentre datasets. As our VideoGAN is a general network architecture, we also evaluate its performance with the CamVid driving video dataset on the cloudy-to-sunny translation task. Comprehensive experiments show that the domain gap could be substantially narrowed down by our VideoGAN.
LT-Net: Label Transfer by Learning Reversible Voxel-wise Correspondence for One-shot Medical Image Segmentation
Mar 20, 2020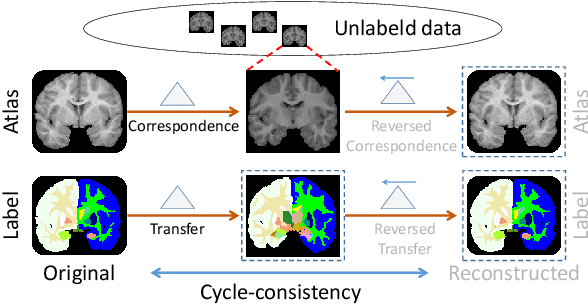
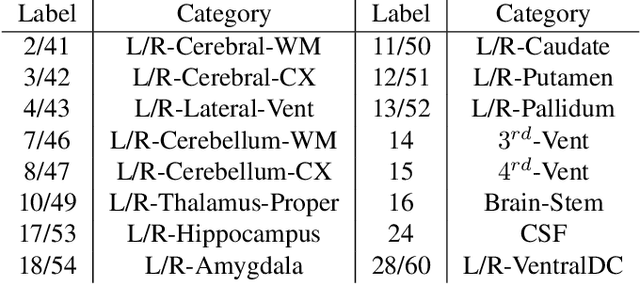
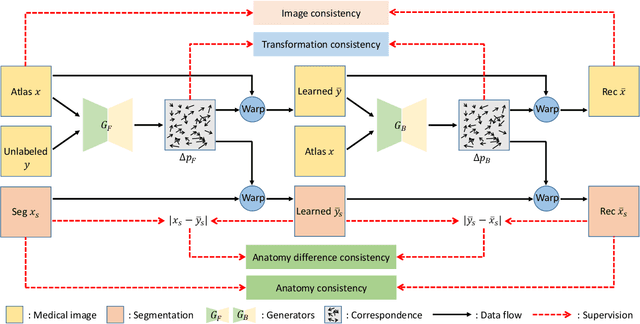
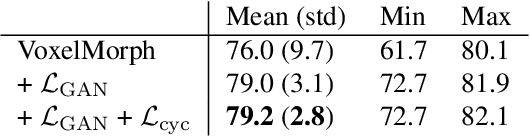
Abstract:We introduce a one-shot segmentation method to alleviate the burden of manual annotation for medical images. The main idea is to treat one-shot segmentation as a classical atlas-based segmentation problem, where voxel-wise correspondence from the atlas to the unlabelled data is learned. Subsequently, segmentation label of the atlas can be transferred to the unlabelled data with the learned correspondence. However, since ground truth correspondence between images is usually unavailable, the learning system must be well-supervised to avoid mode collapse and convergence failure. To overcome this difficulty, we resort to the forward-backward consistency, which is widely used in correspondence problems, and additionally learn the backward correspondences from the warped atlases back to the original atlas. This cycle-correspondence learning design enables a variety of extra, cycle-consistency-based supervision signals to make the training process stable, while also boost the performance. We demonstrate the superiority of our method over both deep learning-based one-shot segmentation methods and a classical multi-atlas segmentation method via thorough experiments.
Quality Control of Neuron Reconstruction Based on Deep Learning
Mar 19, 2020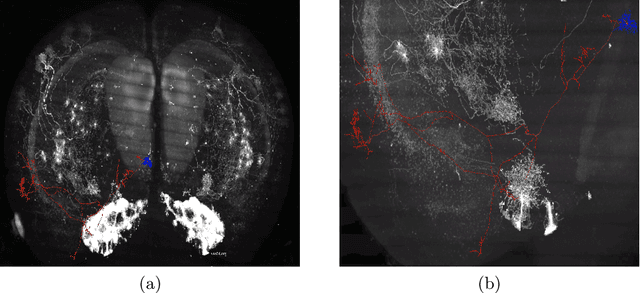
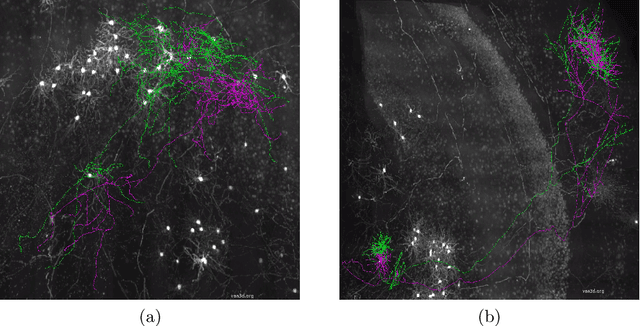
Abstract:Neuron reconstruction is essential to generate exquisite neuron connectivity map for understanding brain function. Despite the significant amount of effect that has been made on automatic reconstruction methods, manual tracing by well-trained human annotators is still necessary. To ensure the quality of reconstructed neurons and provide guidance for annotators to improve their efficiency, we propose a deep learning based quality control method for neuron reconstruction in this paper. By formulating the quality control problem into a binary classification task regarding each single point, the proposed approach overcomes the technical difficulties resulting from the large image size and complex neuron morphology. Not only it provides the evaluation of reconstruction quality, but also can locate exactly where the wrong tracing begins. This work presents one of the first comprehensive studies for whole-brain scale quality control of neuron reconstructions. Experiments on five-fold cross validation with a large dataset demonstrate that the proposed approach can detect 74.7% errors with only 1.4% false alerts.
Identification of primary angle-closure on AS-OCT images with Convolutional Neural Networks
Oct 23, 2019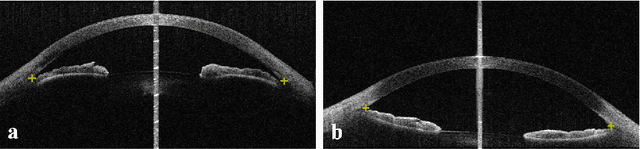
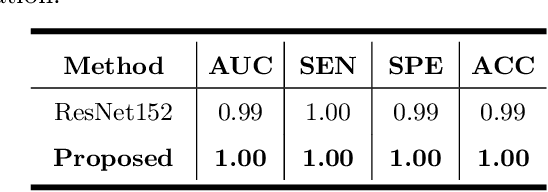
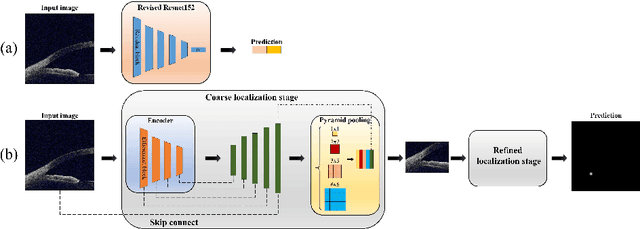
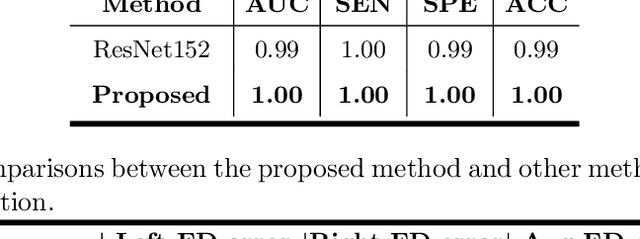
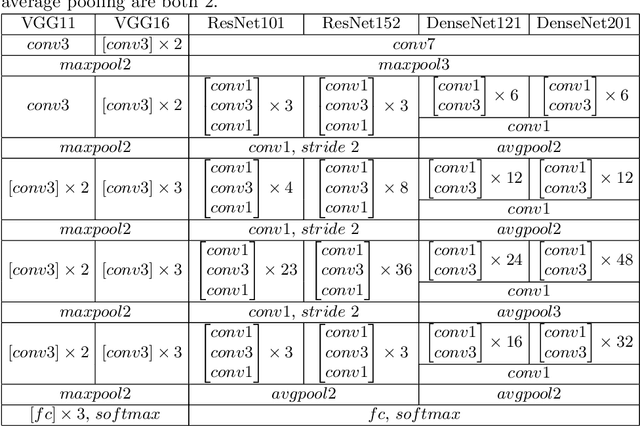
Abstract:Primary angle-closure disease (PACD) is a severe retinal disease, which might cause irreversible vision loss. In clinic, accurate identification of angle-closure and localization of the scleral spur's position on anterior segment optical coherence tomography (AS-OCT) is essential for the diagnosis of PACD. However, manual delineation might confine in low accuracy and low efficiency. In this paper, we propose an efficient and accurate end-to-end architecture for angle-closure classification and scleral spur localization. Specifically, we utilize a revised ResNet152 as our backbone to improve the accuracy of the angle-closure identification. For scleral spur localization, we adopt EfficientNet as encoder because of its powerful feature extraction potential. By combining the skip-connect module and pyramid pooling module, the network is able to collect semantic cues in feature maps from multiple dimensions and scales. Afterward, we propose a novel keypoint registration loss to constrain the model's attention to the intensity and location of the scleral spur area. Several experiments are extensively conducted to evaluate our method on the angle-closure glaucoma evaluation (AGE) Challenge dataset. The results show that our proposed architecture ranks the first place of the classification task on the test dataset and achieves the average Euclidean distance error of 12.00 pixels in the scleral spur localization task.
Self-supervised Feature Learning for 3D Medical Images by Playing a Rubik's Cube
Oct 05, 2019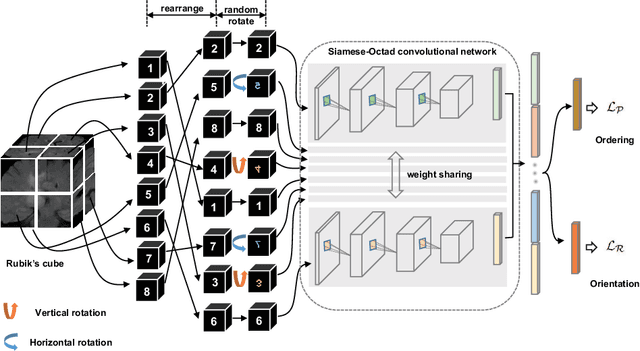
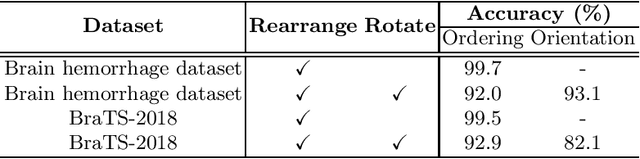
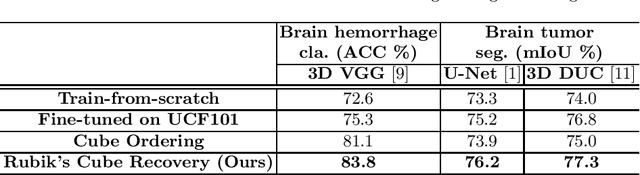
Abstract:Witnessed the development of deep learning, increasing number of studies try to build computer aided diagnosis systems for 3D volumetric medical data. However, as the annotations of 3D medical data are difficult to acquire, the number of annotated 3D medical images is often not enough to well train the deep learning networks. The self-supervised learning deeply exploiting the information of raw data is one of the potential solutions to loose the requirement of training data. In this paper, we propose a self-supervised learning framework for the volumetric medical images. A novel proxy task, i.e., Rubik's cube recovery, is formulated to pre-train 3D neural networks. The proxy task involves two operations, i.e., cube rearrangement and cube rotation, which enforce networks to learn translational and rotational invariant features from raw 3D data. Compared to the train-from-scratch strategy, fine-tuning from the pre-trained network leads to a better accuracy on various tasks, e.g., brain hemorrhage classification and brain tumor segmentation. We show that our self-supervised learning approach can substantially boost the accuracies of 3D deep learning networks on the volumetric medical datasets without using extra data. To our best knowledge, this is the first work focusing on the self-supervised learning of 3D neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge