Kai Ma
School of Electrical Engineering, Yanshan University, Qinhuangdao, China
Domain Adaptation Meets Zero-Shot Learning: An Annotation-Efficient Approach to Multi-Modality Medical Image Segmentation
Mar 19, 2022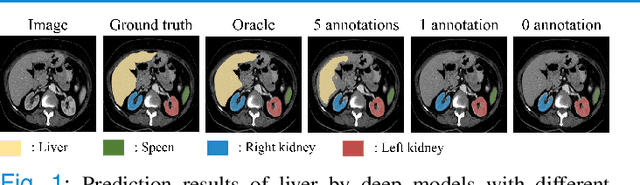
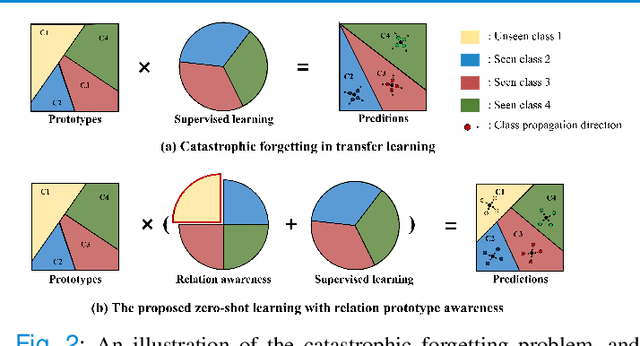
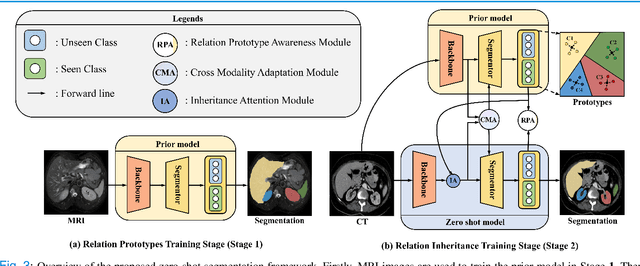
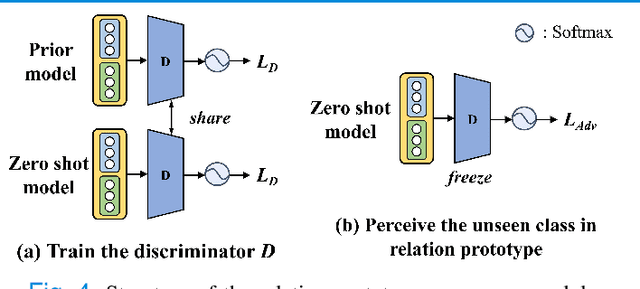
Abstract:Due to the lack of properly annotated medical data, exploring the generalization capability of the deep model is becoming a public concern. Zero-shot learning (ZSL) has emerged in recent years to equip the deep model with the ability to recognize unseen classes. However, existing studies mainly focus on natural images, which utilize linguistic models to extract auxiliary information for ZSL. It is impractical to apply the natural image ZSL solutions directly to medical images, since the medical terminology is very domain-specific, and it is not easy to acquire linguistic models for the medical terminology. In this work, we propose a new paradigm of ZSL specifically for medical images utilizing cross-modality information. We make three main contributions with the proposed paradigm. First, we extract the prior knowledge about the segmentation targets, called relation prototypes, from the prior model and then propose a cross-modality adaptation module to inherit the prototypes to the zero-shot model. Second, we propose a relation prototype awareness module to make the zero-shot model aware of information contained in the prototypes. Last but not least, we develop an inheritance attention module to recalibrate the relation prototypes to enhance the inheritance process. The proposed framework is evaluated on two public cross-modality datasets including a cardiac dataset and an abdominal dataset. Extensive experiments show that the proposed framework significantly outperforms the state of the arts.
Conquering Data Variations in Resolution: A Slice-Aware Multi-Branch Decoder Network
Mar 07, 2022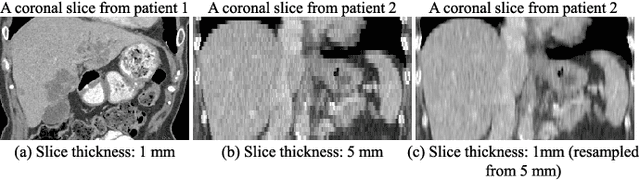
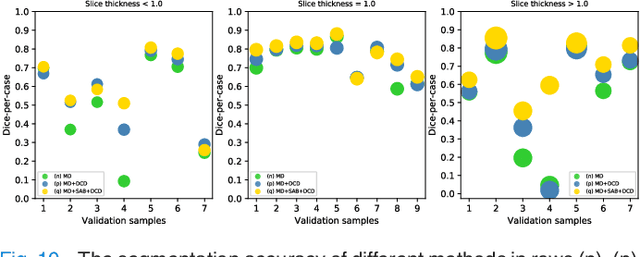
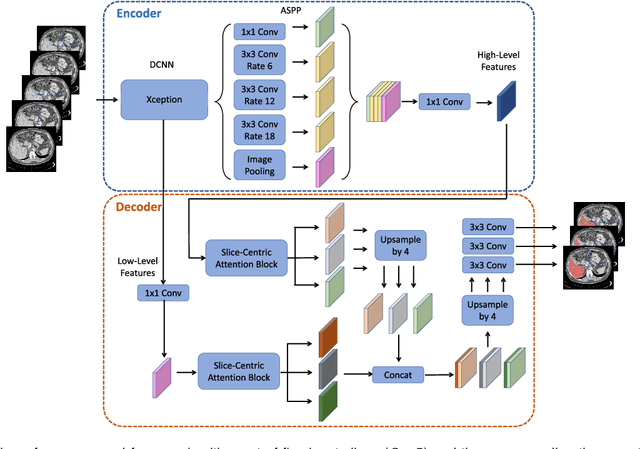
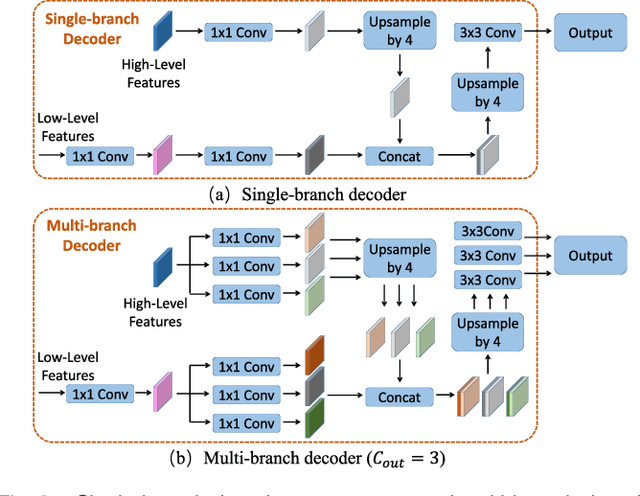
Abstract:Fully convolutional neural networks have made promising progress in joint liver and liver tumor segmentation. Instead of following the debates over 2D versus 3D networks (for example, pursuing the balance between large-scale 2D pretraining and 3D context), in this paper, we novelly identify the wide variation in the ratio between intra- and inter-slice resolutions as a crucial obstacle to the performance. To tackle the mismatch between the intra- and inter-slice information, we propose a slice-aware 2.5D network that emphasizes extracting discriminative features utilizing not only in-plane semantics but also out-of-plane coherence for each separate slice. Specifically, we present a slice-wise multi-input multi-output architecture to instantiate such a design paradigm, which contains a Multi-Branch Decoder (MD) with a Slice-centric Attention Block (SAB) for learning slice-specific features and a Densely Connected Dice (DCD) loss to regularize the inter-slice predictions to be coherent and continuous. Based on the aforementioned innovations, we achieve state-of-the-art results on the MICCAI 2017 Liver Tumor Segmentation (LiTS) dataset. Besides, we also test our model on the ISBI 2019 Segmentation of THoracic Organs at Risk (SegTHOR) dataset, and the result proves the robustness and generalizability of the proposed method in other segmentation tasks.
Simultaneous Alignment and Surface Regression Using Hybrid 2D-3D Networks for 3D Coherent Layer Segmentation of Retina OCT Images
Mar 04, 2022

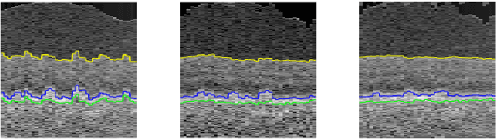
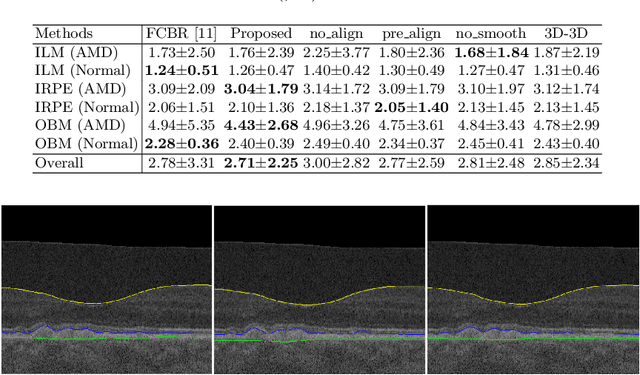
Abstract:Automated surface segmentation of retinal layer is important and challenging in analyzing optical coherence tomography (OCT). Recently, many deep learning based methods have been developed for this task and yield remarkable performance. However, due to large spatial gap and potential mismatch between the B-scans of OCT data, all of them are based on 2D segmentation of individual B-scans, which may loss the continuity information across the B-scans. In addition, 3D surface of the retina layers can provide more diagnostic information, which is crucial in quantitative image analysis. In this study, a novel framework based on hybrid 2D-3D convolutional neural networks (CNNs) is proposed to obtain continuous 3D retinal layer surfaces from OCT. The 2D features of individual B-scans are extracted by an encoder consisting of 2D convolutions. These 2D features are then used to produce the alignment displacement field and layer segmentation by two 3D decoders, which are coupled via a spatial transformer module. The entire framework is trained end-to-end. To the best of our knowledge, this is the first study that attempts 3D retinal layer segmentation in volumetric OCT images based on CNNs. Experiments on a publicly available dataset show that our framework achieves superior results to state-of-the-art 2D methods in terms of both layer segmentation accuracy and cross-B-scan 3D continuity, thus offering more clinical values than previous works.
Bidirectional Pricing and Demand Response for Nanogrids with HVAC Systems
Mar 01, 2022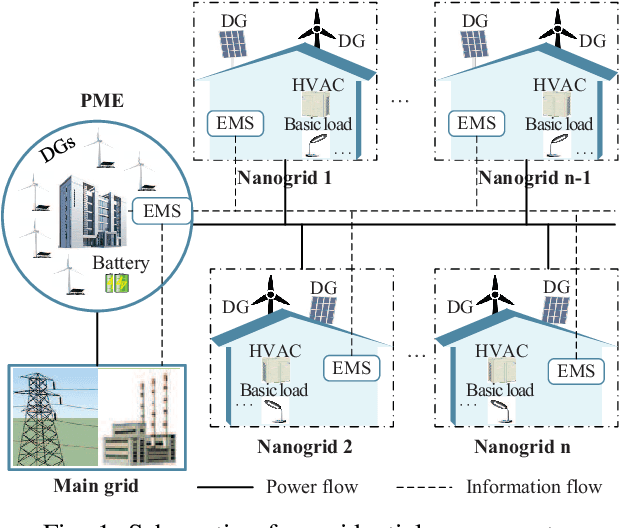
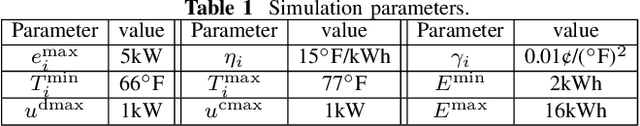
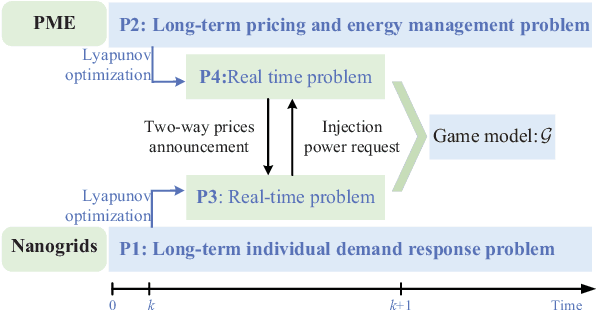
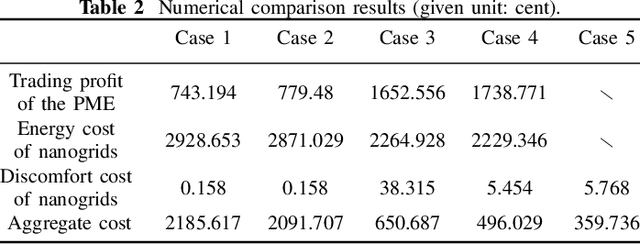
Abstract:Owing to the fluctuant renewable generation and power demand, the energy surplus or deficit in each nanogrid is embodied differently across time. To stimulate local renewable energy consumption and minimize the long-term energy cost, some issues still remain to be explored: when and how the energy demand and bidirectional trading prices are scheduled considering personal comfort preferences and environmental factors. For this purpose, the demand response and two-way pricing problems concurrently for nanogrids and a public monitoring entity (PME) are studied with exploiting the large potential thermal elastic ability of heating, ventilation and air-conditioning (HVAC) units. Different from nanogrids, in terms of minimizing time-average costs, PME aims to set reasonable prices and optimize profits by trading with nanogrids and the main grid bi-directionally. In particular, such bilevel energy management problem is formulated as a stochastic form in a long-term horizon. Since there are uncertain system parameters, time-coupled queue constraints and the interplay of bilevel decision-making, it is challenging to solve the formulated problems. To this end, we derive a form of relaxation based on Lyapunov optimization technique to make the energy management problem tractable without forecasting the related system parameters. The transaction between nanogrids and PME is captured by a one-leader and multi-follower Stackelberg game framework. Then, theoretical analysis of the existence and uniqueness of Stackelberg equilibrium (SE) is developed based on the proposed game property. Following that, we devise an optimization algorithm to reach the SE with less information exchange. Numerical experiments validate the effectiveness of the proposed approach.
Asynchronous Decentralized Federated Learning for Collaborative Fault Diagnosis of PV Stations
Feb 28, 2022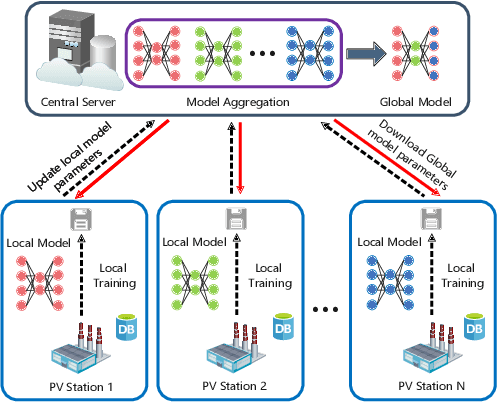
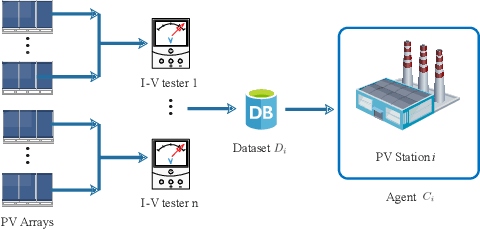
Abstract:Due to the different losses caused by various photovoltaic (PV) array faults, accurate diagnosis of fault types is becoming increasingly important. Compared with a single one, multiple PV stations collect sufficient fault samples, but their data is not allowed to be shared directly due to potential conflicts of interest. Therefore, federated learning can be exploited to train a collaborative fault diagnosis model. However, the modeling efficiency is seriously affected by the model update mechanism since each PV station has a different computing capability and amount of data. Moreover, for the safe and stable operation of the PV system, the robustness of collaborative modeling must be guaranteed rather than simply being processed on a central server. To address these challenges, a novel asynchronous decentralized federated learning (ADFL) framework is proposed. Each PV station not only trains its local model but also participates in collaborative fault diagnosis by exchanging model parameters to improve the generalization without losing accuracy. The global model is aggregated distributedly to avoid central node failure. By designing the asynchronous update scheme, the communication overhead and training time are greatly reduced. Both the experiments and numerical simulations are carried out to verify the effectiveness of the proposed method.
Label Propagation for Annotation-Efficient Nuclei Segmentation from Pathology Images
Feb 16, 2022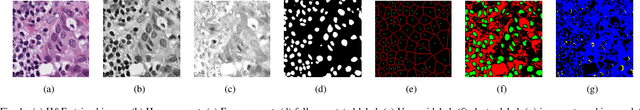
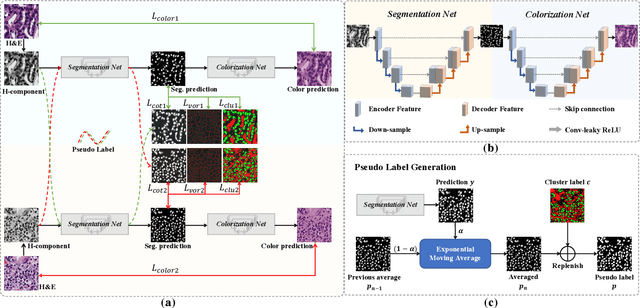
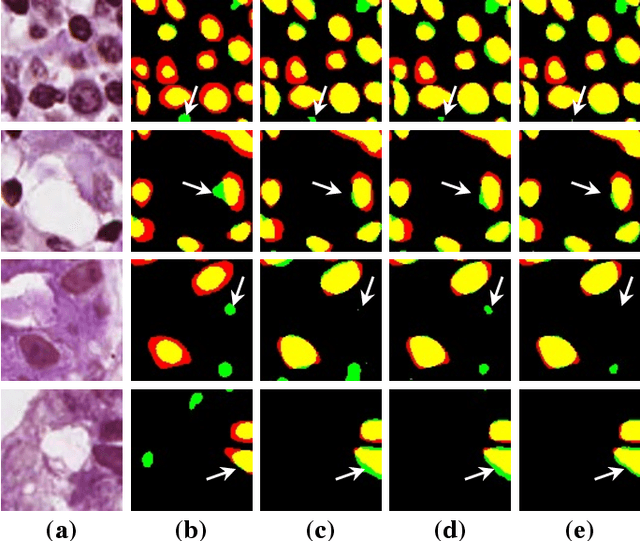
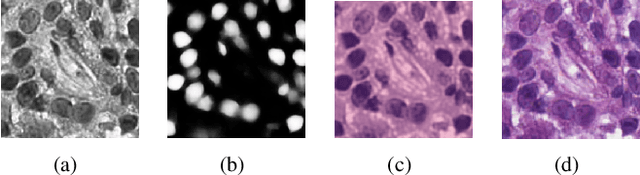
Abstract:Nuclei segmentation is a crucial task for whole slide image analysis in digital pathology. Generally, the segmentation performance of fully-supervised learning heavily depends on the amount and quality of the annotated data. However, it is time-consuming and expensive for professional pathologists to provide accurate pixel-level ground truth, while it is much easier to get coarse labels such as point annotations. In this paper, we propose a weakly-supervised learning method for nuclei segmentation that only requires point annotations for training. The proposed method achieves label propagation in a coarse-to-fine manner as follows. First, coarse pixel-level labels are derived from the point annotations based on the Voronoi diagram and the k-means clustering method to avoid overfitting. Second, a co-training strategy with an exponential moving average method is designed to refine the incomplete supervision of the coarse labels. Third, a self-supervised visual representation learning method is tailored for nuclei segmentation of pathology images that transforms the hematoxylin component images into the H\&E stained images to gain better understanding of the relationship between the nuclei and cytoplasm. We comprehensively evaluate the proposed method using two public datasets. Both visual and quantitative results demonstrate the superiority of our method to the state-of-the-art methods, and its competitive performance compared to the fully-supervised methods. The source codes for implementing the experiments will be released after acceptance.
Revisiting Experience Replay: Continual Learning by Adaptively Tuning Task-wise Relationship
Jan 06, 2022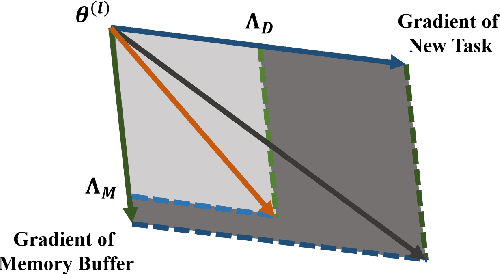
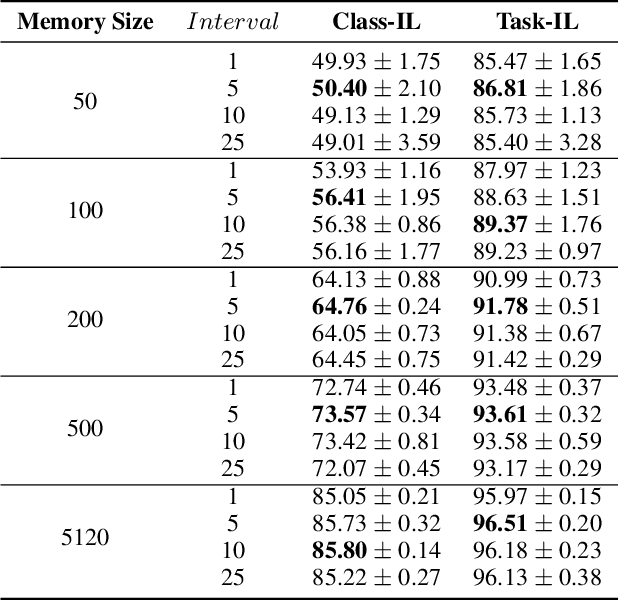
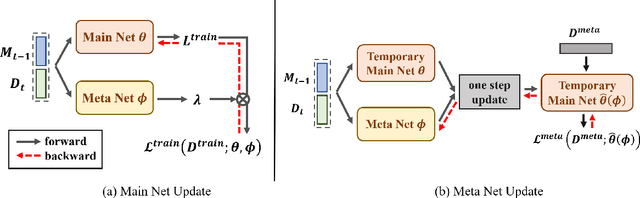
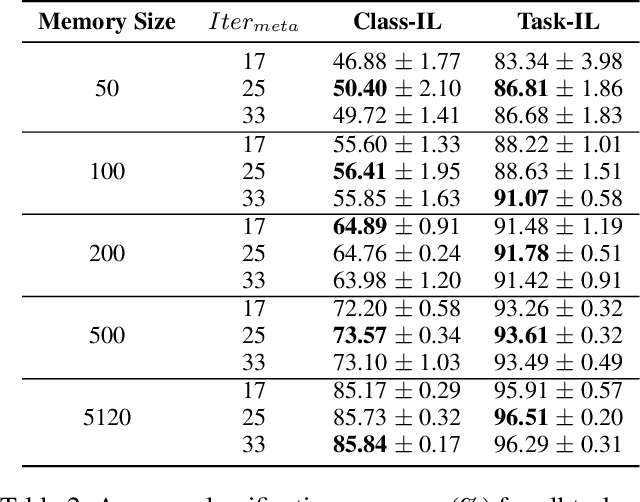
Abstract:Continual learning requires models to learn new tasks while maintaining previously learned knowledge. Various algorithms have been proposed to address this real challenge. Till now, rehearsal-based methods, such as experience replay, have achieved state-of-the-art performance. These approaches save a small part of the data of the past tasks as a memory buffer to prevent models from forgetting previously learned knowledge. However, most of them treat every new task equally, i.e., fixed the hyperparameters of the framework while learning different new tasks. Such a setting lacks the consideration of the relationship/similarity between past and new tasks. For example, the previous knowledge/features learned from dogs are more beneficial for the identification of cats (new task), compared to those learned from buses. In this regard, we propose a meta learning algorithm based on bi-level optimization to adaptively tune the relationship between the knowledge extracted from the past and new tasks. Therefore, the model can find an appropriate direction of gradient during continual learning and avoid the serious overfitting problem on memory buffer. Extensive experiments are conducted on three publicly available datasets (i.e., CIFAR-10, CIFAR-100, and Tiny ImageNet). The experimental results demonstrate that the proposed method can consistently improve the performance of all baselines.
Alleviating Noisy-label Effects in Image Classification via Probability Transition Matrix
Oct 19, 2021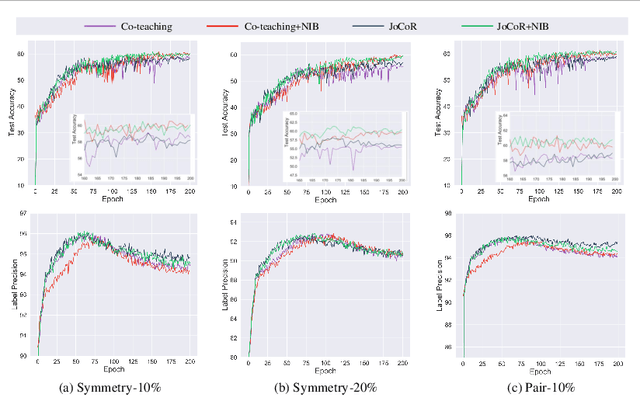
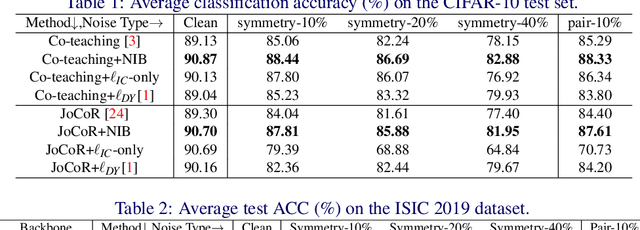
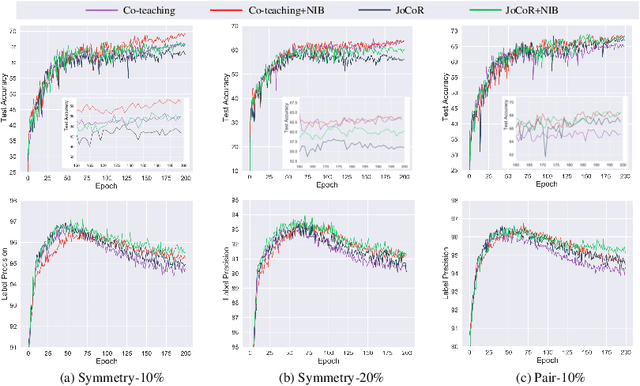
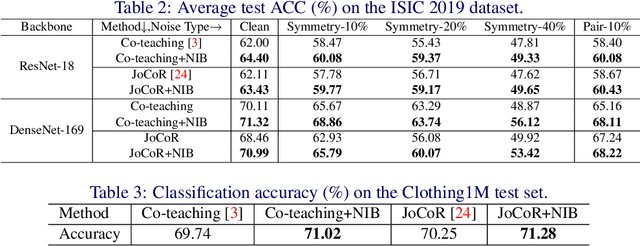
Abstract:Deep-learning-based image classification frameworks often suffer from the noisy label problem caused by the inter-observer variation. Recent studies employed learning-to-learn paradigms (e.g., Co-teaching and JoCoR) to filter the samples with noisy labels from the training set. However, most of them use a simple cross-entropy loss as the criterion for noisy label identification. The hard samples, which are beneficial for classifier learning, are often mistakenly treated as noises in such a setting since both the hard samples and ones with noisy labels lead to a relatively larger loss value than the easy cases. In this paper, we propose a plugin module, namely noise ignoring block (NIB), consisting of a probability transition matrix and an inter-class correlation (IC) loss, to separate the hard samples from the mislabeled ones, and further boost the accuracy of image classification network trained with noisy labels. Concretely, our IC loss is calculated as Kullback-Leibler divergence between the network prediction and the accumulative soft label generated by the probability transition matrix. Such that, with the lower value of IC loss, the hard cases can be easily distinguished from mislabeled cases. Extensive experiments are conducted on natural and medical image datasets (CIFAR-10 and ISIC 2019). The experimental results show that our NIB module consistently improves the performances of the state-of-the-art robust training methods.
A Unified Framework for Generalized Low-Shot Medical Image Segmentation with Scarce Data
Oct 18, 2021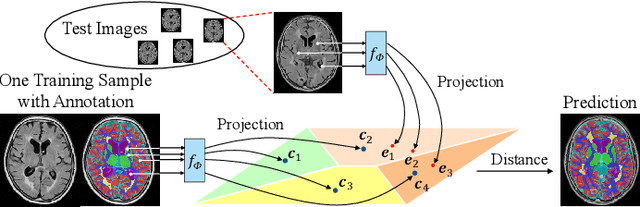
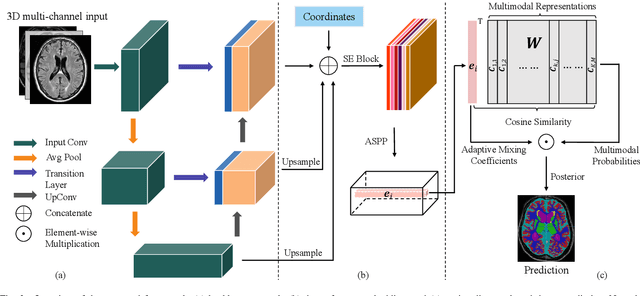
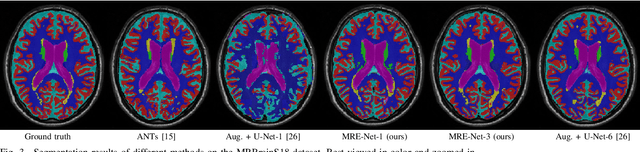
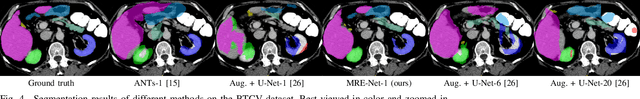
Abstract:Medical image segmentation has achieved remarkable advancements using deep neural networks (DNNs). However, DNNs often need big amounts of data and annotations for training, both of which can be difficult and costly to obtain. In this work, we propose a unified framework for generalized low-shot (one- and few-shot) medical image segmentation based on distance metric learning (DML). Unlike most existing methods which only deal with the lack of annotations while assuming abundance of data, our framework works with extreme scarcity of both, which is ideal for rare diseases. Via DML, the framework learns a multimodal mixture representation for each category, and performs dense predictions based on cosine distances between the pixels' deep embeddings and the category representations. The multimodal representations effectively utilize the inter-subject similarities and intraclass variations to overcome overfitting due to extremely limited data. In addition, we propose adaptive mixing coefficients for the multimodal mixture distributions to adaptively emphasize the modes better suited to the current input. The representations are implicitly embedded as weights of the fc layer, such that the cosine distances can be computed efficiently via forward propagation. In our experiments on brain MRI and abdominal CT datasets, the proposed framework achieves superior performances for low-shot segmentation towards standard DNN-based (3D U-Net) and classical registration-based (ANTs) methods, e.g., achieving mean Dice coefficients of 81%/69% for brain tissue/abdominal multiorgan segmentation using a single training sample, as compared to 52%/31% and 72%/35% by the U-Net and ANTs, respectively.
Unsupervised Representation Learning Meets Pseudo-Label Supervised Self-Distillation: A New Approach to Rare Disease Classification
Oct 09, 2021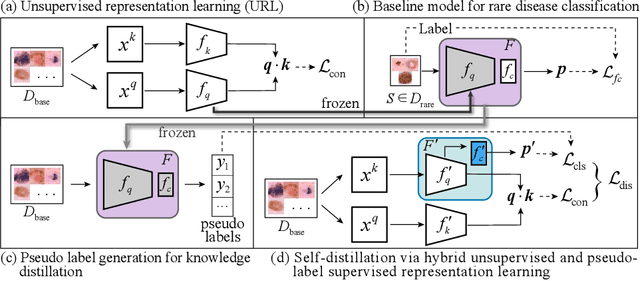
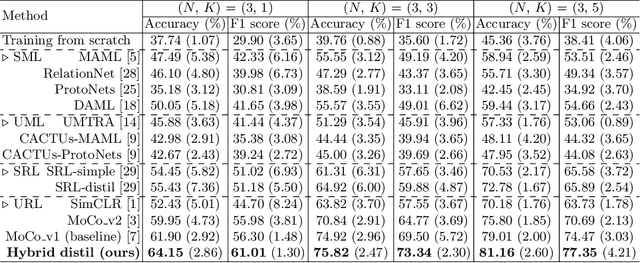
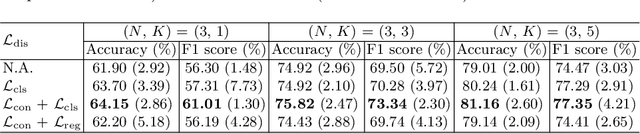
Abstract:Rare diseases are characterized by low prevalence and are often chronically debilitating or life-threatening. Imaging-based classification of rare diseases is challenging due to the severe shortage in training examples. Few-shot learning (FSL) methods tackle this challenge by extracting generalizable prior knowledge from a large base dataset of common diseases and normal controls, and transferring the knowledge to rare diseases. Yet, most existing methods require the base dataset to be labeled and do not make full use of the precious examples of the rare diseases. To this end, we propose in this work a novel hybrid approach to rare disease classification, featuring two key novelties targeted at the above drawbacks. First, we adopt the unsupervised representation learning (URL) based on self-supervising contrastive loss, whereby to eliminate the overhead in labeling the base dataset. Second, we integrate the URL with pseudo-label supervised classification for effective self-distillation of the knowledge about the rare diseases, composing a hybrid approach taking advantages of both unsupervised and (pseudo-) supervised learning on the base dataset. Experimental results on classification of rare skin lesions show that our hybrid approach substantially outperforms existing FSL methods (including those using fully supervised base dataset) for rare disease classification via effective integration of the URL and pseudo-label driven self-distillation, thus establishing a new state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge