Domain-adversarial neural networks to address the appearance variability of histopathology images
Jul 19, 2017Maxime W. Lafarge, Josien P. W. Pluim, Koen A. J. Eppenhof, Pim Moeskops, Mitko Veta
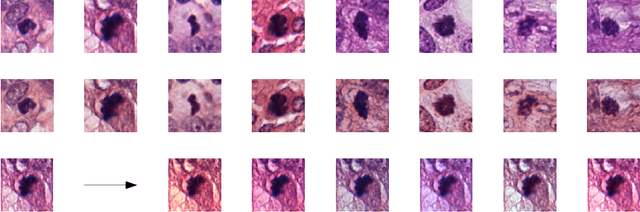
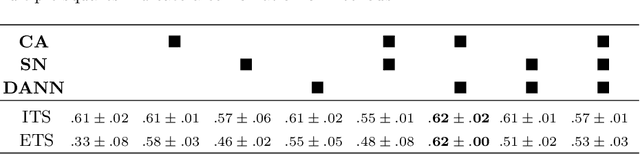
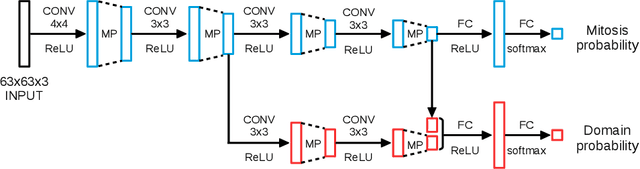
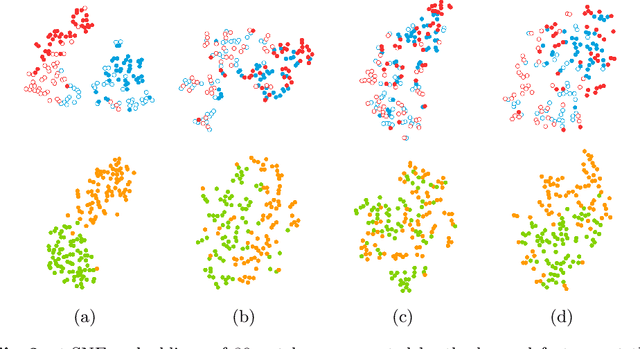
Preparing and scanning histopathology slides consists of several steps, each with a multitude of parameters. The parameters can vary between pathology labs and within the same lab over time, resulting in significant variability of the tissue appearance that hampers the generalization of automatic image analysis methods. Typically, this is addressed with ad-hoc approaches such as staining normalization that aim to reduce the appearance variability. In this paper, we propose a systematic solution based on domain-adversarial neural networks. We hypothesize that removing the domain information from the model representation leads to better generalization. We tested our hypothesis for the problem of mitosis detection in breast cancer histopathology images and made a comparative analysis with two other approaches. We show that combining color augmentation with domain-adversarial training is a better alternative than standard approaches to improve the generalization of deep learning methods.
Adversarial training and dilated convolutions for brain MRI segmentation
Jul 11, 2017Pim Moeskops, Mitko Veta, Maxime W. Lafarge, Koen A. J. Eppenhof, Josien P. W. Pluim
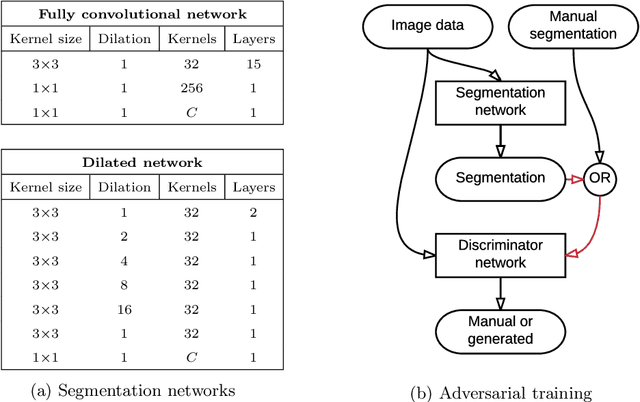
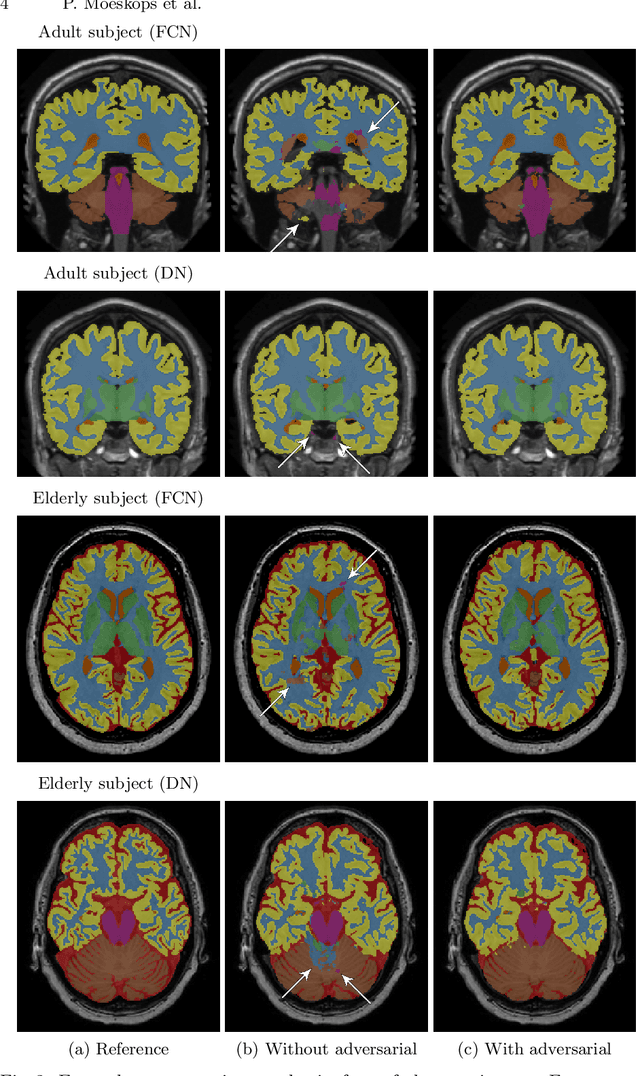
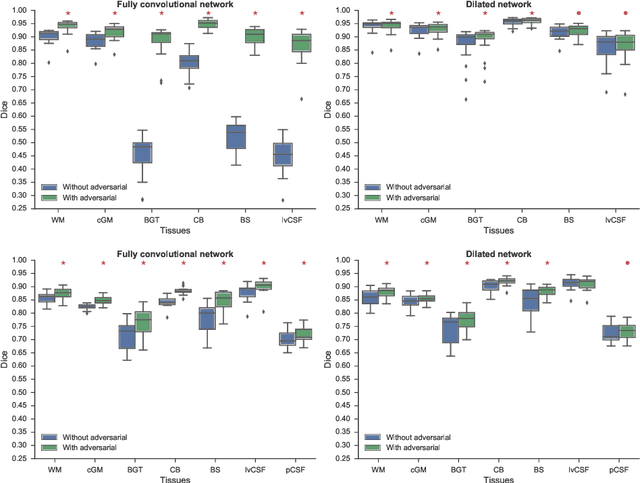
Convolutional neural networks (CNNs) have been applied to various automatic image segmentation tasks in medical image analysis, including brain MRI segmentation. Generative adversarial networks have recently gained popularity because of their power in generating images that are difficult to distinguish from real images. In this study we use an adversarial training approach to improve CNN-based brain MRI segmentation. To this end, we include an additional loss function that motivates the network to generate segmentations that are difficult to distinguish from manual segmentations. During training, this loss function is optimised together with the conventional average per-voxel cross entropy loss. The results show improved segmentation performance using this adversarial training procedure for segmentation of two different sets of images and using two different network architectures, both visually and in terms of Dice coefficients.
Gland Segmentation in Colon Histology Images: The GlaS Challenge Contest
Sep 01, 2016Korsuk Sirinukunwattana, Josien P. W. Pluim, Hao Chen, Xiaojuan Qi, Pheng-Ann Heng, Yun Bo Guo, Li Yang Wang, Bogdan J. Matuszewski, Elia Bruni, Urko Sanchez, Anton Böhm, Olaf Ronneberger, Bassem Ben Cheikh, Daniel Racoceanu, Philipp Kainz, Michael Pfeiffer, Martin Urschler, David R. J. Snead, Nasir M. Rajpoot
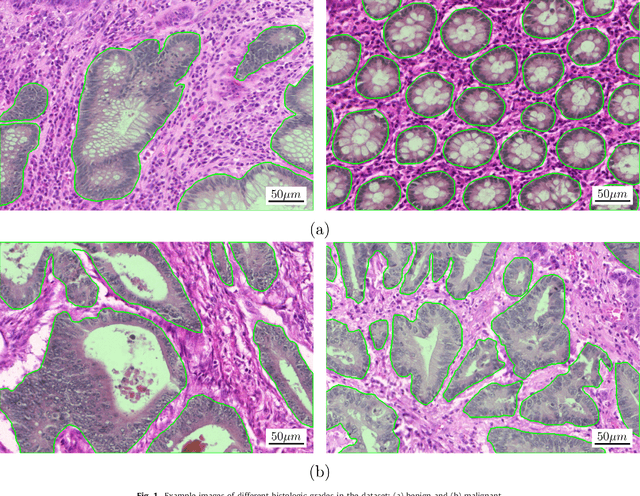
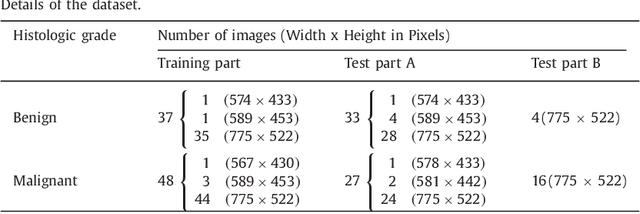
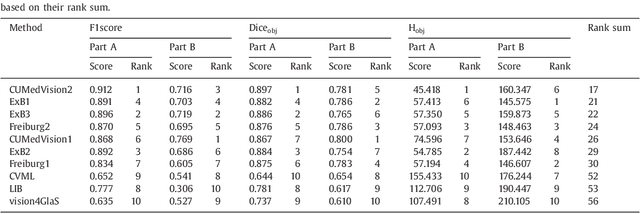
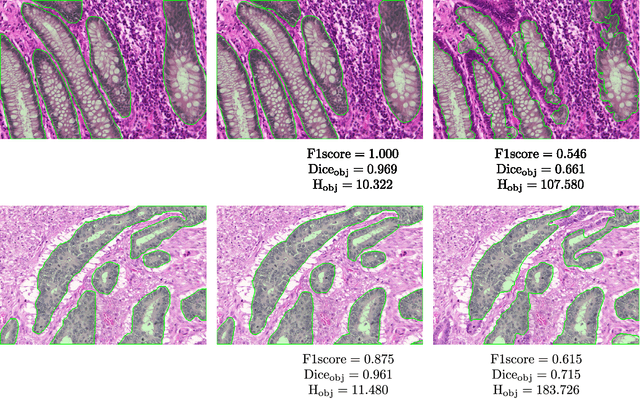
Colorectal adenocarcinoma originating in intestinal glandular structures is the most common form of colon cancer. In clinical practice, the morphology of intestinal glands, including architectural appearance and glandular formation, is used by pathologists to inform prognosis and plan the treatment of individual patients. However, achieving good inter-observer as well as intra-observer reproducibility of cancer grading is still a major challenge in modern pathology. An automated approach which quantifies the morphology of glands is a solution to the problem. This paper provides an overview to the Gland Segmentation in Colon Histology Images Challenge Contest (GlaS) held at MICCAI'2015. Details of the challenge, including organization, dataset and evaluation criteria, are presented, along with the method descriptions and evaluation results from the top performing methods.
Cutting out the middleman: measuring nuclear area in histopathology slides without segmentation
Jun 20, 2016Mitko Veta, Paul J. van Diest, Josien P. W. Pluim
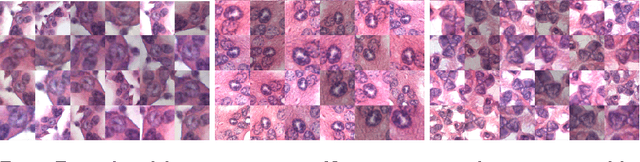
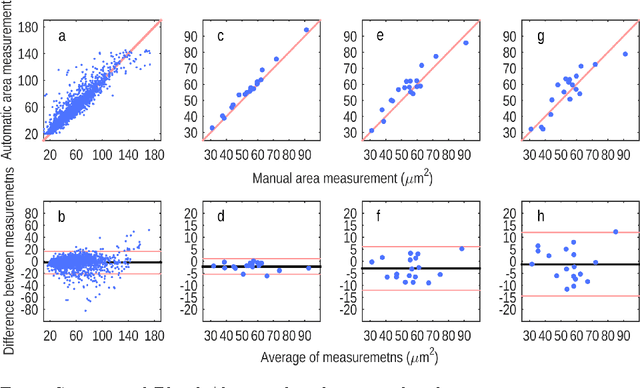
The size of nuclei in histological preparations from excised breast tumors is predictive of patient outcome (large nuclei indicate poor outcome). Pathologists take into account nuclear size when performing breast cancer grading. In addition, the mean nuclear area (MNA) has been shown to have independent prognostic value. The straightforward approach to measuring nuclear size is by performing nuclei segmentation. We hypothesize that given an image of a tumor region with known nuclei locations, the area of the individual nuclei and region statistics such as the MNA can be reliably computed directly from the image data by employing a machine learning model, without the intermediate step of nuclei segmentation. Towards this goal, we train a deep convolutional neural network model that is applied locally at each nucleus location, and can reliably measure the area of the individual nuclei and the MNA. Furthermore, we show how such an approach can be extended to perform combined nuclei detection and measurement, which is reminiscent of granulometry.
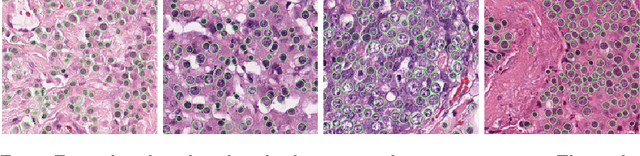
Assessment of algorithms for mitosis detection in breast cancer histopathology images
Nov 21, 2014Mitko Veta, Paul J. van Diest, Stefan M. Willems, Haibo Wang, Anant Madabhushi, Angel Cruz-Roa, Fabio Gonzalez, Anders B. L. Larsen, Jacob S. Vestergaard, Anders B. Dahl, Dan C. Cireşan, Jürgen Schmidhuber, Alessandro Giusti, Luca M. Gambardella, F. Boray Tek, Thomas Walter, Ching-Wei Wang, Satoshi Kondo, Bogdan J. Matuszewski, Frederic Precioso, Violet Snell, Josef Kittler, Teofilo E. de Campos, Adnan M. Khan, Nasir M. Rajpoot, Evdokia Arkoumani, Miangela M. Lacle, Max A. Viergever, Josien P. W. Pluim
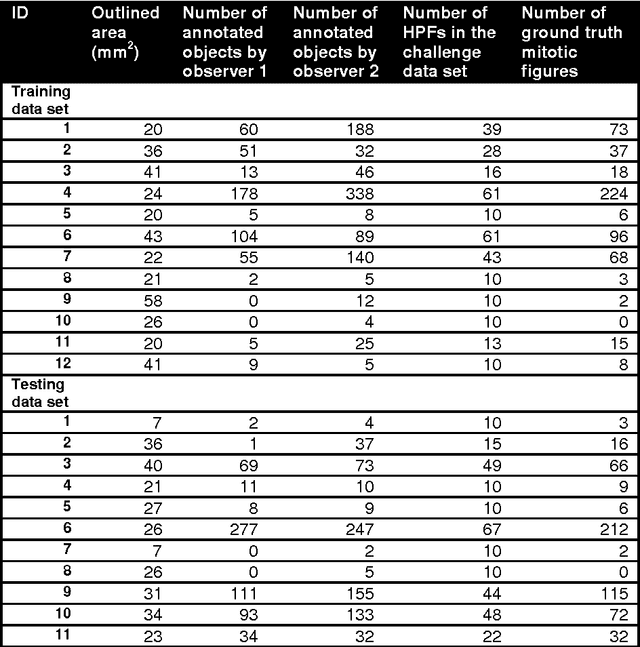
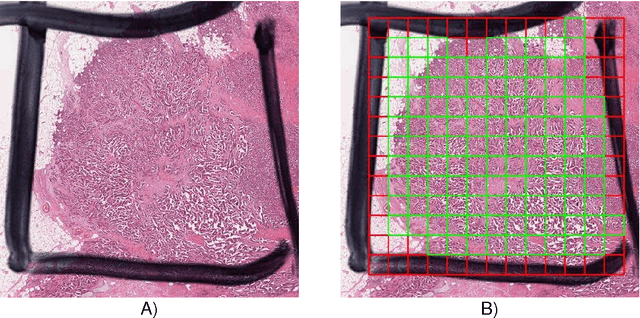
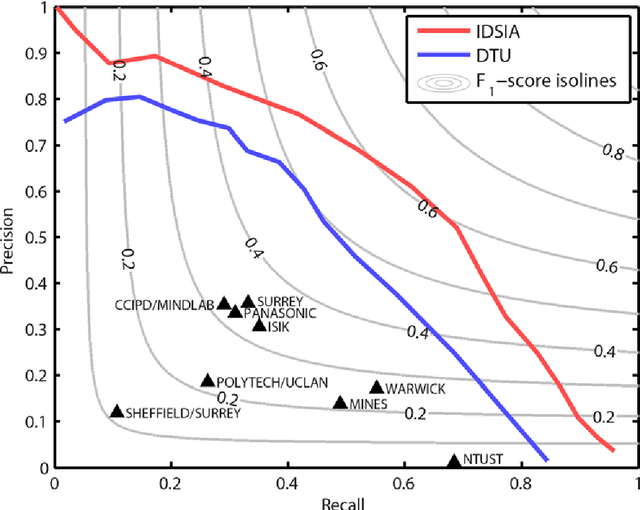
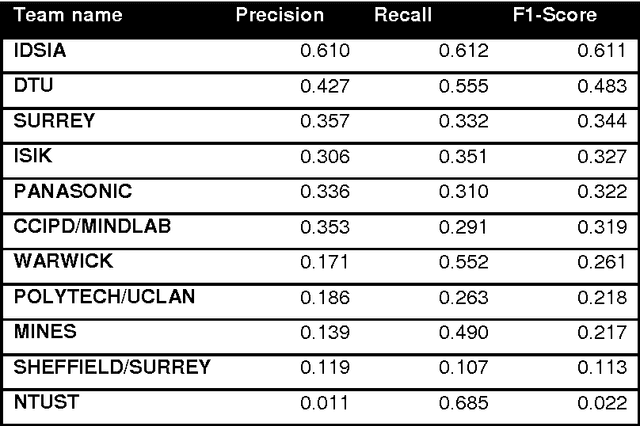
The proliferative activity of breast tumors, which is routinely estimated by counting of mitotic figures in hematoxylin and eosin stained histology sections, is considered to be one of the most important prognostic markers. However, mitosis counting is laborious, subjective and may suffer from low inter-observer agreement. With the wider acceptance of whole slide images in pathology labs, automatic image analysis has been proposed as a potential solution for these issues. In this paper, the results from the Assessment of Mitosis Detection Algorithms 2013 (AMIDA13) challenge are described. The challenge was based on a data set consisting of 12 training and 11 testing subjects, with more than one thousand annotated mitotic figures by multiple observers. Short descriptions and results from the evaluation of eleven methods are presented. The top performing method has an error rate that is comparable to the inter-observer agreement among pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge