Jonas Richiardi
From 100,000+ images to winning the first brain MRI foundation model challenges: Sharing lessons and models
Jan 19, 2026Abstract:Developing Foundation Models for medical image analysis is essential to overcome the unique challenges of radiological tasks. The first challenges of this kind for 3D brain MRI, SSL3D and FOMO25, were held at MICCAI 2025. Our solution ranked first in tracks of both contests. It relies on a U-Net CNN architecture combined with strategies leveraging anatomical priors and neuroimaging domain knowledge. Notably, our models trained 1-2 orders of magnitude faster and were 10 times smaller than competing transformer-based approaches. Models are available here: https://github.com/jbanusco/BrainFM4Challenges.
Spatiotemporal graph neural process for reconstruction, extrapolation, and classification of cardiac trajectories
Sep 16, 2025



Abstract:We present a probabilistic framework for modeling structured spatiotemporal dynamics from sparse observations, focusing on cardiac motion. Our approach integrates neural ordinary differential equations (NODEs), graph neural networks (GNNs), and neural processes into a unified model that captures uncertainty, temporal continuity, and anatomical structure. We represent dynamic systems as spatiotemporal multiplex graphs and model their latent trajectories using a GNN-parameterized vector field. Given the sparse context observations at node and edge levels, the model infers a distribution over latent initial states and control variables, enabling both interpolation and extrapolation of trajectories. We validate the method on three synthetic dynamical systems (coupled pendulum, Lorenz attractor, and Kuramoto oscillators) and two real-world cardiac imaging datasets - ACDC (N=150) and UK Biobank (N=526) - demonstrating accurate reconstruction, extrapolation, and disease classification capabilities. The model accurately reconstructs trajectories and extrapolates future cardiac cycles from a single observed cycle. It achieves state-of-the-art results on the ACDC classification task (up to 99% accuracy), and detects atrial fibrillation in UK Biobank subjects with competitive performance (up to 67% accuracy). This work introduces a flexible approach for analyzing cardiac motion and offers a foundation for graph-based learning in structured biomedical spatiotemporal time-series data.
Fast refacing of MR images with a generative neural network lowers re-identification risk and preserves volumetric consistency
May 26, 2023Abstract:With the rise of open data, identifiability of individuals based on 3D renderings obtained from routine structural magnetic resonance imaging (MRI) scans of the head has become a growing privacy concern. To protect subject privacy, several algorithms have been developed to de-identify imaging data using blurring, defacing or refacing. Completely removing facial structures provides the best re-identification protection but can significantly impact post-processing steps, like brain morphometry. As an alternative, refacing methods that replace individual facial structures with generic templates have a lower effect on the geometry and intensity distribution of original scans, and are able to provide more consistent post-processing results by the price of higher re-identification risk and computational complexity. In the current study, we propose a novel method for anonymised face generation for defaced 3D T1-weighted scans based on a 3D conditional generative adversarial network. To evaluate the performance of the proposed de-identification tool, a comparative study was conducted between several existing defacing and refacing tools, with two different segmentation algorithms (FAST and Morphobox). The aim was to evaluate (i) impact on brain morphometry reproducibility, (ii) re-identification risk, (iii) balance between (i) and (ii), and (iv) the processing time. The proposed method takes 9 seconds for face generation and is suitable for recovering consistent post-processing results after defacing.
Transfer learning with weak labels from radiology reports: application to glioma change detection
Oct 18, 2022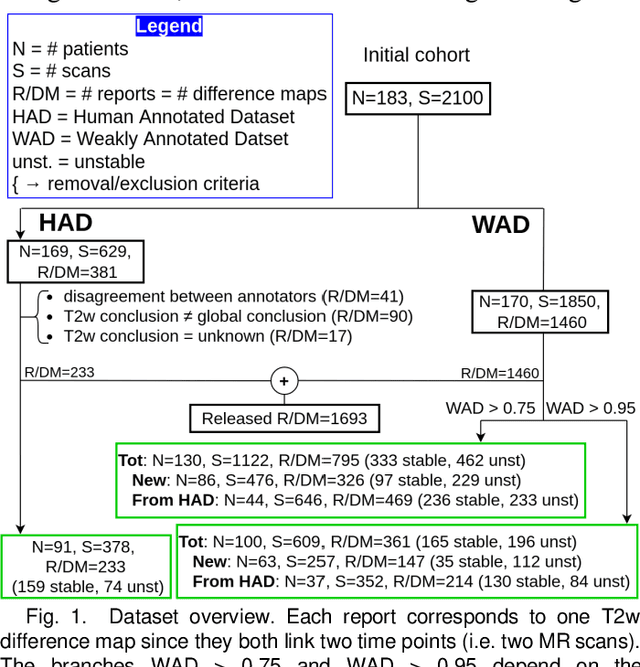
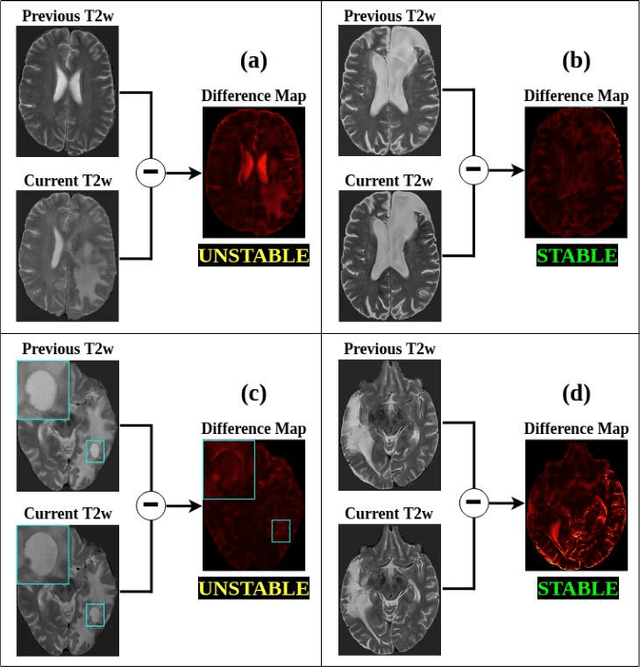
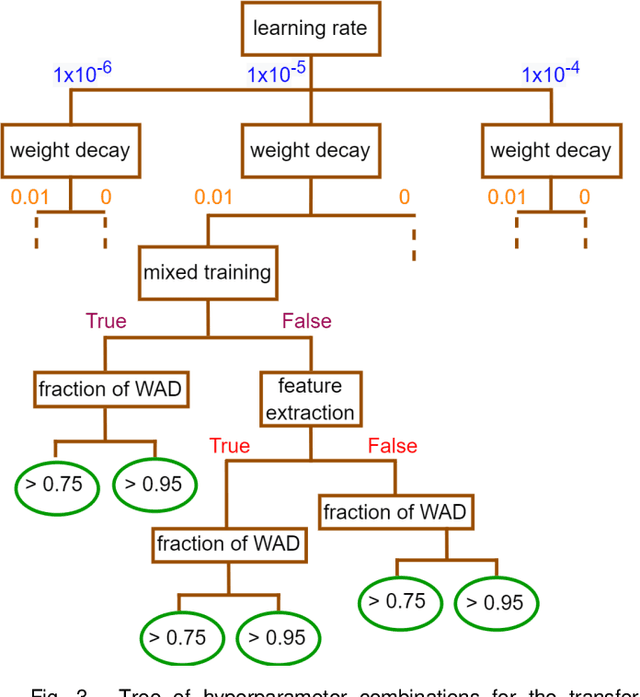
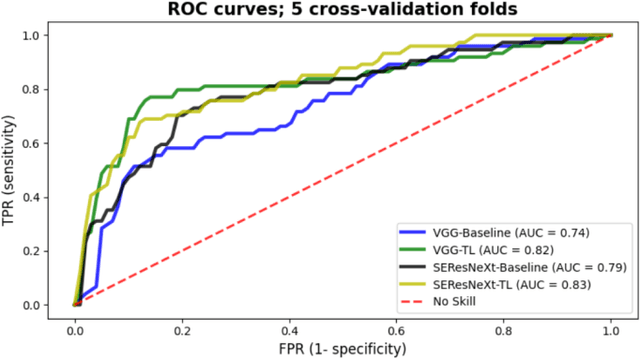
Abstract:Creating large annotated datasets represents a major bottleneck for the development of deep learning models in radiology. To overcome this, we propose a combined use of weak labels (imprecise, but fast-to-create annotations) and Transfer Learning (TL). Specifically, we explore inductive TL, where source and target domains are identical, but tasks are different due to a label shift: our target labels are created manually by three radiologists, whereas the source weak labels are generated automatically from textual radiology reports. We frame knowledge transfer as hyperparameter optimization, thus avoiding heuristic choices that are frequent in related works. We investigate the relationship between model size and TL, comparing a low-capacity VGG with a higher-capacity SEResNeXt. The task that we address is change detection in follow-up glioma imaging: we extracted 1693 T2-weighted magnetic resonance imaging difference maps from 183 patients, and classified them into stable or unstable according to tumor evolution. Weak labeling allowed us to increase dataset size more than 3-fold, and improve VGG classification results from 75% to 82% Area Under the ROC Curve (AUC) (p=0.04). Mixed training from scratch led to higher performance than fine-tuning or feature extraction. To assess generalizability, we also ran inference on an open dataset (BraTS-2015: 15 patients, 51 difference maps), reaching up to 76% AUC. Overall, results suggest that medical imaging problems may benefit from smaller models and different TL strategies with respect to computer vision problems, and that report-generated weak labels are effective in improving model performances. Code, in-house dataset and BraTS labels are released.
The Multiscenario Multienvironment BioSecure Multimodal Database (BMDB)
Nov 17, 2021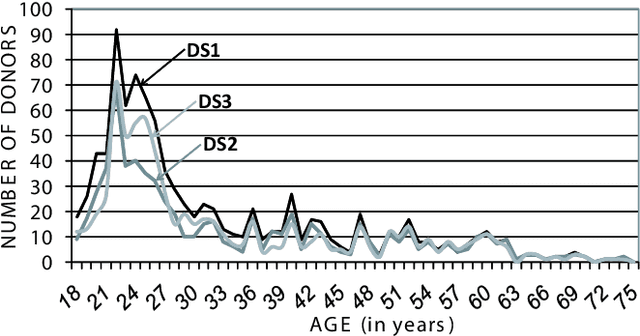

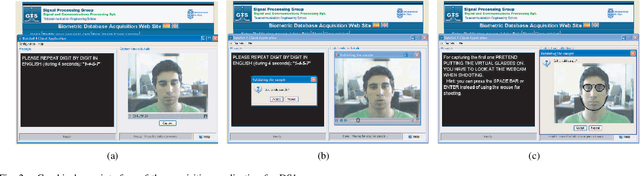
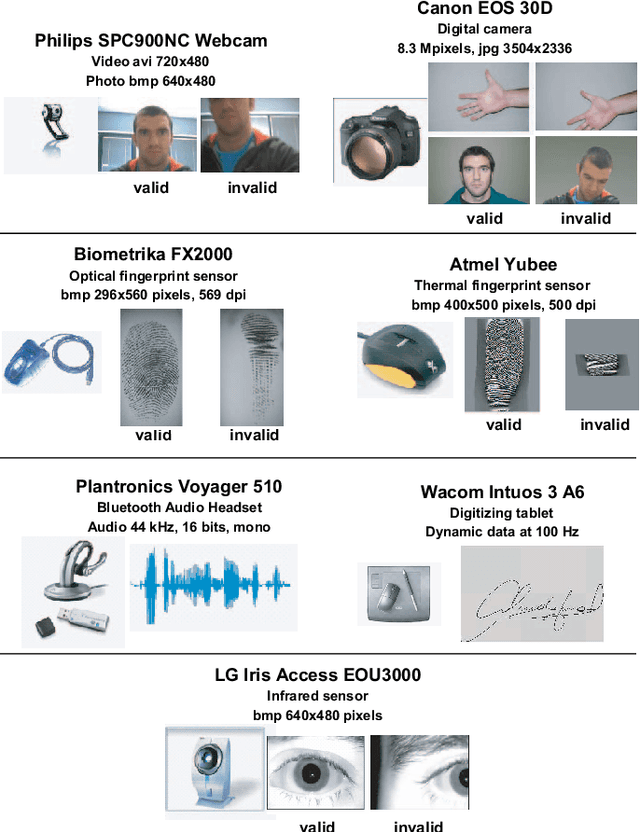
Abstract:A new multimodal biometric database designed and acquired within the framework of the European BioSecure Network of Excellence is presented. It is comprised of more than 600 individuals acquired simultaneously in three scenarios: 1) over the Internet, 2) in an office environment with desktop PC, and 3) in indoor/outdoor environments with mobile portable hardware. The three scenarios include a common part of audio/video data. Also, signature and fingerprint data have been acquired both with desktop PC and mobile portable hardware. Additionally, hand and iris data were acquired in the second scenario using desktop PC. Acquisition has been conducted by 11 European institutions. Additional features of the BioSecure Multimodal Database (BMDB) are: two acquisition sessions, several sensors in certain modalities, balanced gender and age distributions, multimodal realistic scenarios with simple and quick tasks per modality, cross-European diversity, availability of demographic data, and compatibility with other multimodal databases. The novel acquisition conditions of the BMDB allow us to perform new challenging research and evaluation of either monomodal or multimodal biometric systems, as in the recent BioSecure Multimodal Evaluation campaign. A description of this campaign including baseline results of individual modalities from the new database is also given. The database is expected to be available for research purposes through the BioSecure Association during 2008
Weak labels and anatomical knowledge: making deep learning practical for intracranial aneurysm detection in TOF-MRA
Mar 10, 2021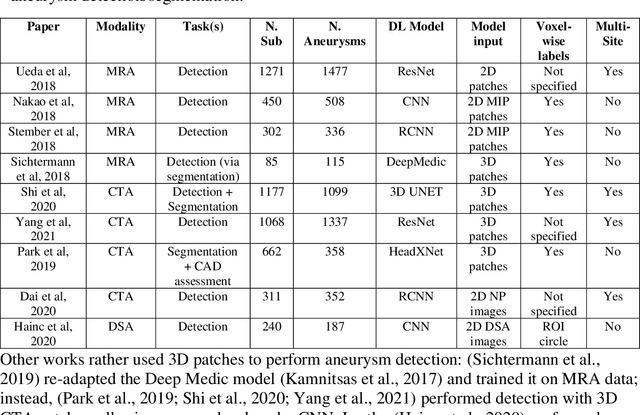
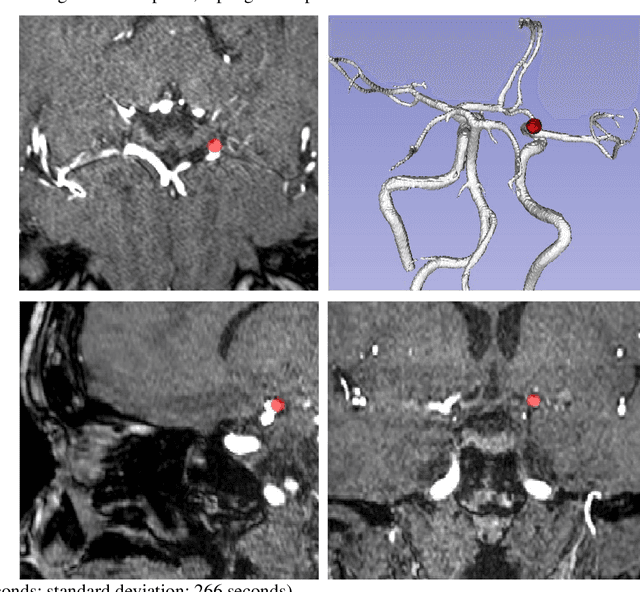
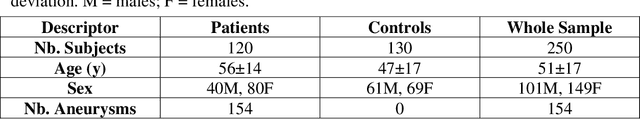
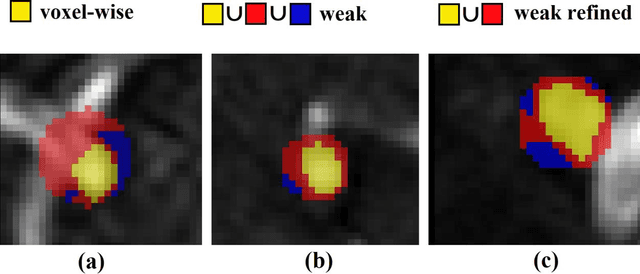
Abstract:Supervised segmentation algorithms yield state-of-the-art results for automated anomaly detection. However, these models require voxel-wise labels which are time-consuming to draw for medical experts. An interesting alternative to voxel-wise annotations is the use of weak labels: these can be coarse or oversized annotations that are less precise, but considerably faster to create. In this work, we address the task of brain aneurysm detection by developing a fully automated, deep neural network that is trained utilizing oversized weak labels. Furthermore, since aneurysms mainly occur in specific anatomical locations, we build our model leveraging the underlying anatomy of the brain vasculature both during training and inference. We apply our model to 250 subjects (120 patients, 130 controls) who underwent Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) and presented a total of 154 aneurysms. To assess the robustness of the algorithm, we participated in a MICCAI challenge for TOF-MRA data (93 patients, 20 controls, 125 aneurysms) which allowed us to obtain results also for subjects coming from a different institution. Our network achieves an average sensitivity of 77% on our in-house data, with a mean False Positive (FP) rate of 0.72 per patient. Instead, on the challenge data, we attain a sensitivity of 59% with a mean FP rate of 1.18, ranking in 7th/14 position for detection and in 4th/11 for segmentation on the open leaderboard. When computing detection performances with respect to aneurysms' risk of rupture, we found no statistical difference between two risk groups (p = 0.12), although the sensitivity for dangerous aneurysms was higher (78%). Our approach suggests that clinically useful sensitivity can be achieved using weak labels and exploiting prior anatomical knowledge; this expands the feasibility of deep learning studies to hospitals that have limited time and data.
An anatomically-informed 3D CNN for brain aneurysm classification with weak labels
Nov 27, 2020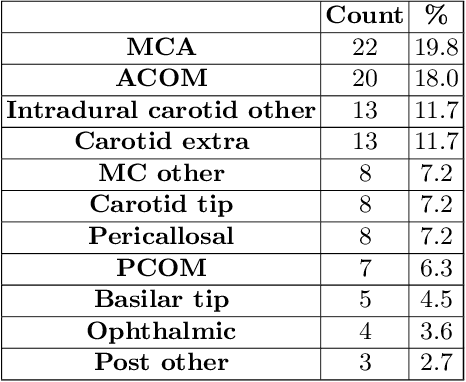
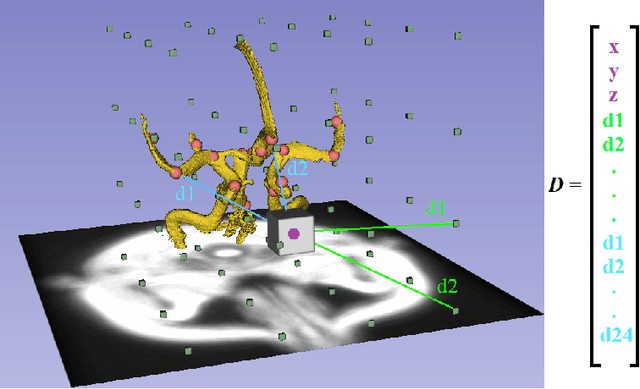
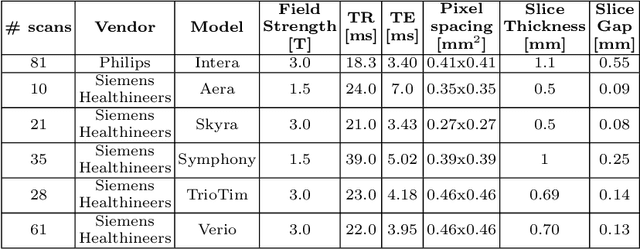
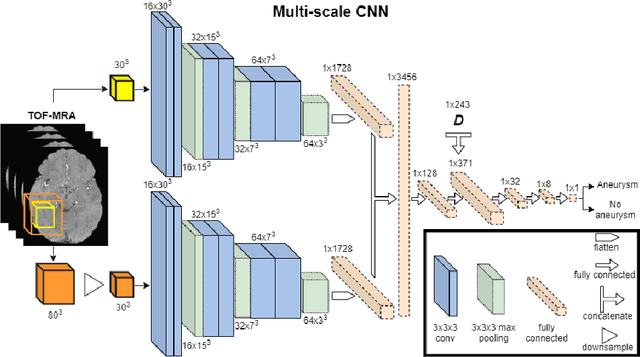
Abstract:A commonly adopted approach to carry out detection tasks in medical imaging is to rely on an initial segmentation. However, this approach strongly depends on voxel-wise annotations which are repetitive and time-consuming to draw for medical experts. An interesting alternative to voxel-wise masks are so-called "weak" labels: these can either be coarse or oversized annotations that are less precise, but noticeably faster to create. In this work, we address the task of brain aneurysm detection as a patch-wise binary classification with weak labels, in contrast to related studies that rather use supervised segmentation methods and voxel-wise delineations. Our approach comes with the non-trivial challenge of the data set creation: as for most focal diseases, anomalous patches (with aneurysm) are outnumbered by those showing no anomaly, and the two classes usually have different spatial distributions. To tackle this frequent scenario of inherently imbalanced, spatially skewed data sets, we propose a novel, anatomically-driven approach by using a multi-scale and multi-input 3D Convolutional Neural Network (CNN). We apply our model to 214 subjects (83 patients, 131 controls) who underwent Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) and presented a total of 111 unruptured cerebral aneurysms. We compare two strategies for negative patch sampling that have an increasing level of difficulty for the network and we show how this choice can strongly affect the results. To assess whether the added spatial information helps improving performances, we compare our anatomically-informed CNN with a baseline, spatially-agnostic CNN. When considering the more realistic and challenging scenario including vessel-like negative patches, the former model attains the highest classification results (accuracy$\simeq$95\%, AUROC$\simeq$0.95, AUPR$\simeq$0.71), thus outperforming the baseline.
Shallow vs deep learning architectures for white matter lesion segmentation in the early stages of multiple sclerosis
Sep 10, 2018
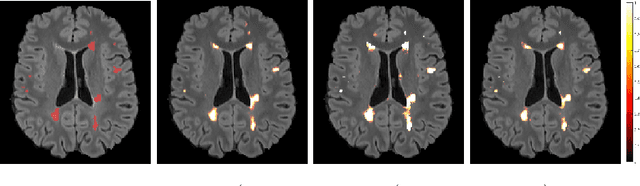
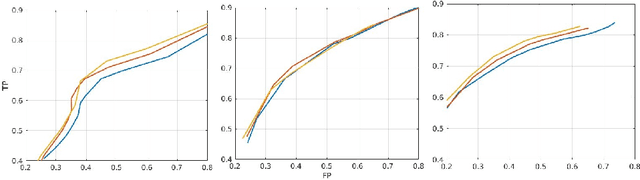
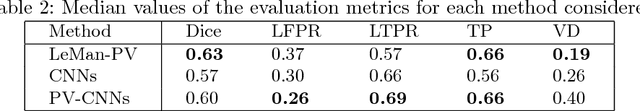
Abstract:In this work, we present a comparison of a shallow and a deep learning architecture for the automated segmentation of white matter lesions in MR images of multiple sclerosis patients. In particular, we train and test both methods on early stage disease patients, to verify their performance in challenging conditions, more similar to a clinical setting than what is typically provided in multiple sclerosis segmentation challenges. Furthermore, we evaluate a prototype naive combination of the two methods, which refines the final segmentation. All methods were trained on 32 patients, and the evaluation was performed on a pure test set of 73 cases. Results show low lesion-wise false positives (30%) for the deep learning architecture, whereas the shallow architecture yields the best Dice coefficient (63%) and volume difference (19%). Combining both shallow and deep architectures further improves the lesion-wise metrics (69% and 26% lesion-wise true and false positive rate, respectively).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge