Augustin C. Ogier
Spatiotemporal graph neural process for reconstruction, extrapolation, and classification of cardiac trajectories
Sep 16, 2025



Abstract:We present a probabilistic framework for modeling structured spatiotemporal dynamics from sparse observations, focusing on cardiac motion. Our approach integrates neural ordinary differential equations (NODEs), graph neural networks (GNNs), and neural processes into a unified model that captures uncertainty, temporal continuity, and anatomical structure. We represent dynamic systems as spatiotemporal multiplex graphs and model their latent trajectories using a GNN-parameterized vector field. Given the sparse context observations at node and edge levels, the model infers a distribution over latent initial states and control variables, enabling both interpolation and extrapolation of trajectories. We validate the method on three synthetic dynamical systems (coupled pendulum, Lorenz attractor, and Kuramoto oscillators) and two real-world cardiac imaging datasets - ACDC (N=150) and UK Biobank (N=526) - demonstrating accurate reconstruction, extrapolation, and disease classification capabilities. The model accurately reconstructs trajectories and extrapolates future cardiac cycles from a single observed cycle. It achieves state-of-the-art results on the ACDC classification task (up to 99% accuracy), and detects atrial fibrillation in UK Biobank subjects with competitive performance (up to 67% accuracy). This work introduces a flexible approach for analyzing cardiac motion and offers a foundation for graph-based learning in structured biomedical spatiotemporal time-series data.
Three-dimensional reconstruction and characterization of bladder deformations
Jan 18, 2023



Abstract:Background and Objective: Pelvic floor disorders are prevalent diseases and patient care remains difficult as the dynamics of the pelvic floor remains poorly known. So far, only 2D dynamic observations of straining exercises at excretion are available in the clinics and the understanding of three-dimensional pelvic organs mechanical defects is not yet achievable. In this context, we proposed a complete methodology for the 3D representation of the non-reversible bladder deformations during exercises, directly combined with synthesized 3D representation of the location of the highest strain areas on the organ surface. Methods: Novel image segmentation and registration approaches have been combined with three geometrical configurations of up-to-date rapid dynamic multi-slices MRI acquisition for the reconstruction of real-time dynamic bladder volumes. Results: For the first time, we proposed real-time 3D deformation fields of the bladder under strain from in-bore forced breathing exercises. The potential of our method was assessed on eight control subjects undergoing forced breathing exercises. We obtained average volume deviation of the reconstructed dynamic volume of bladders around 2.5\% and high registration accuracy with mean distance values of 0.4 $\pm$ 0.3 mm and Hausdorff distance values of 2.2 $\pm$ 1.1 mm. Conclusions: Immediately transferable to the clinics with rapid acquisitions, the proposed framework represents a real advance in the field of pelvic floor disorders as it provides, for the first time, a proper 3D+t spatial tracking of bladder non-reversible deformations. This work is intended to be extended to patients with cavities filling and excretion to better characterize the degree of severity of pelvic floor pathologies for diagnostic assistance or in preoperative surgical planning.
A new geodesic-based feature for characterization of 3D shapes: application to soft tissue organ temporal deformations
Mar 18, 2020

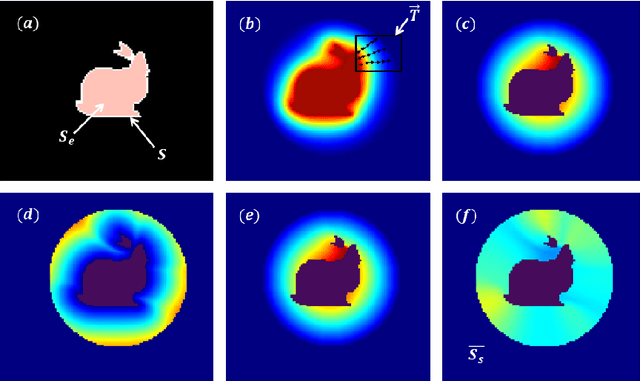
Abstract:In this paper, we propose a method for characterizing 3D shapes from point clouds and we show a direct application on a study of organ temporal deformations. As an example, we characterize the behavior of a bladder during a forced respiratory motion with a reduced number of 3D surface points: first, a set of equidistant points representing the vertices of quadrilateral mesh for the surface in the first time frame are tracked throughout a long dynamic MRI sequence using a Large Deformation Diffeomorphic Metric Mapping (LDDMM) framework. Second, a novel geometric feature which is invariant to scaling and rotation is proposed for characterizing the temporal organ deformations by employing an Eulerian Partial Differential Equations (PDEs) methodology. We demonstrate the robustness of our feature on both synthetic 3D shapes and realistic dynamic MRI data portraying the bladder deformation during forced respiratory motions. Promising results are obtained, showing that the proposed feature may be useful for several computer vision applications such as medical imaging, aerodynamics and robotics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge