Patric Hagmann
Department of Radiology, Lausanne University Hospital and University of Lausanne
Comparing and Scaling fMRI Features for Brain-Behavior Prediction
Jul 28, 2025Abstract:Predicting behavioral variables from neuroimaging modalities such as magnetic resonance imaging (MRI) has the potential to allow the development of neuroimaging biomarkers of mental and neurological disorders. A crucial processing step to this aim is the extraction of suitable features. These can differ in how well they predict the target of interest, and how this prediction scales with sample size and scan time. Here, we compare nine feature subtypes extracted from resting-state functional MRI recordings for behavior prediction, ranging from regional measures of functional activity to functional connectivity (FC) and metrics derived with graph signal processing (GSP), a principled approach for the extraction of structure-informed functional features. We study 979 subjects from the Human Connectome Project Young Adult dataset, predicting summary scores for mental health, cognition, processing speed, and substance use, as well as age and sex. The scaling properties of the features are investigated for different combinations of sample size and scan time. FC comes out as the best feature for predicting cognition, age, and sex. Graph power spectral density is the second best for predicting cognition and age, while for sex, variability-based features show potential as well. When predicting sex, the low-pass graph filtered coupled FC slightly outperforms the simple FC variant. None of the other targets were predicted significantly. The scaling results point to higher performance reserves for the better-performing features. They also indicate that it is important to balance sample size and scan time when acquiring data for prediction studies. The results confirm FC as a robust feature for behavior prediction, but also show the potential of GSP and variability-based measures. We discuss the implications for future prediction studies in terms of strategies for acquisition and sample composition.
Predicting Cognition from fMRI:A Comparative Study of Graph, Transformer, and Kernel Models Across Task and Rest Conditions
Jul 28, 2025Abstract:Predicting cognition from neuroimaging data in healthy individuals offers insights into the neural mechanisms underlying cognitive abilities, with potential applications in precision medicine and early detection of neurological and psychiatric conditions. This study systematically benchmarked classical machine learning (Kernel Ridge Regression (KRR)) and advanced deep learning (DL) models (Graph Neural Networks (GNN) and Transformer-GNN (TGNN)) for cognitive prediction using Resting-state (RS), Working Memory, and Language task fMRI data from the Human Connectome Project Young Adult dataset. Our results, based on R2 scores, Pearson correlation coefficient, and mean absolute error, revealed that task-based fMRI, eliciting neural responses directly tied to cognition, outperformed RS fMRI in predicting cognitive behavior. Among the methods compared, a GNN combining structural connectivity (SC) and functional connectivity (FC) consistently achieved the highest performance across all fMRI modalities; however, its advantage over KRR using FC alone was not statistically significant. The TGNN, designed to model temporal dynamics with SC as a prior, performed competitively with FC-based approaches for task-fMRI but struggled with RS data, where its performance aligned with the lower-performing GNN that directly used fMRI time-series data as node features. These findings emphasize the importance of selecting appropriate model architectures and feature representations to fully leverage the spatial and temporal richness of neuroimaging data. This study highlights the potential of multimodal graph-aware DL models to combine SC and FC for cognitive prediction, as well as the promise of Transformer-based approaches for capturing temporal dynamics. By providing a comprehensive comparison of models, this work serves as a guide for advancing brain-behavior modeling using fMRI, SC and DL.
Transfer learning with weak labels from radiology reports: application to glioma change detection
Oct 18, 2022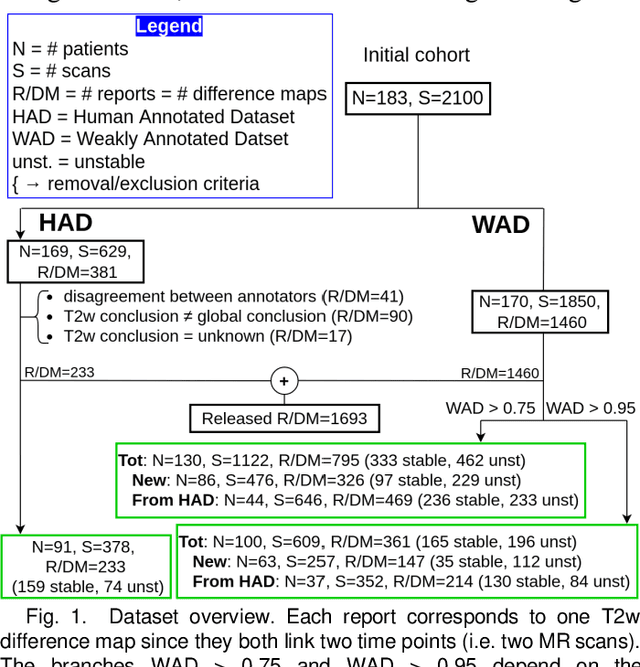
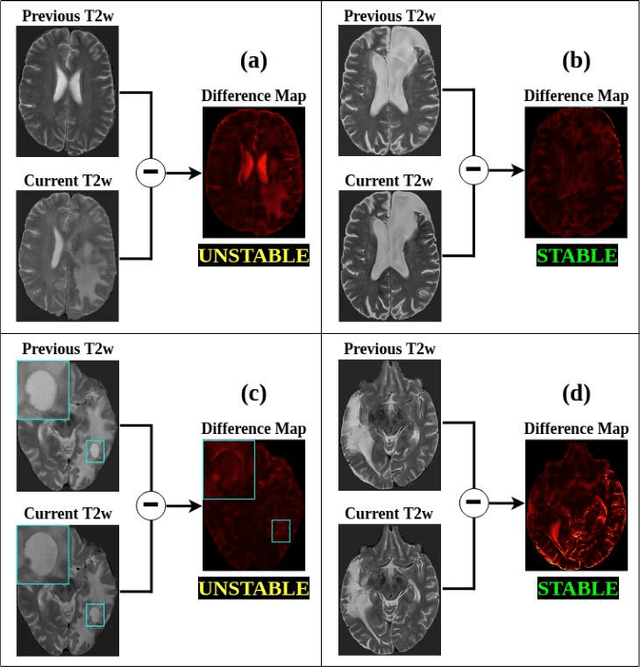
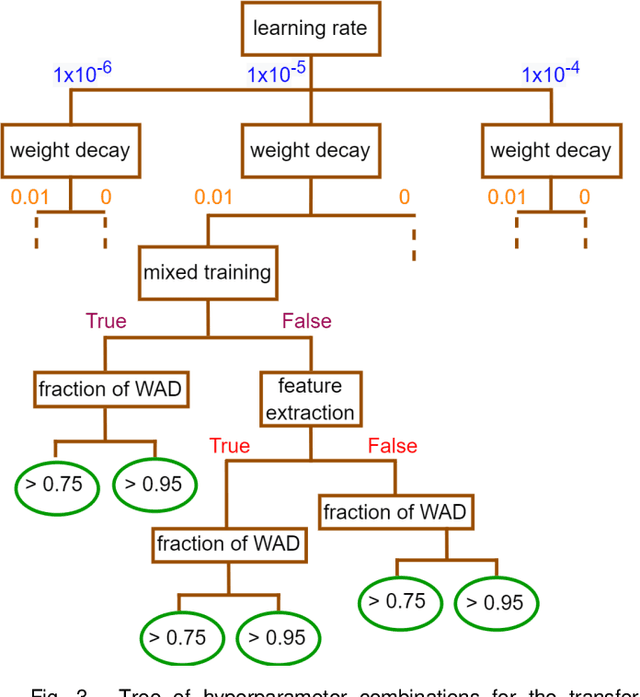
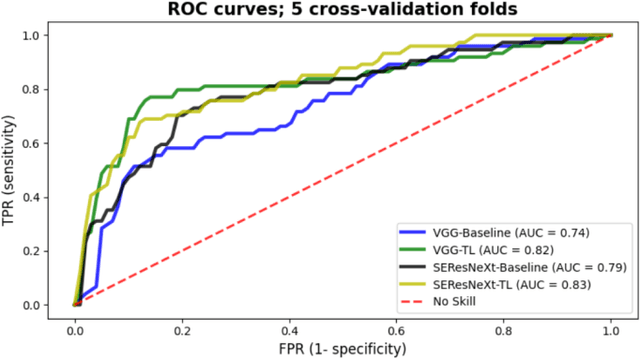
Abstract:Creating large annotated datasets represents a major bottleneck for the development of deep learning models in radiology. To overcome this, we propose a combined use of weak labels (imprecise, but fast-to-create annotations) and Transfer Learning (TL). Specifically, we explore inductive TL, where source and target domains are identical, but tasks are different due to a label shift: our target labels are created manually by three radiologists, whereas the source weak labels are generated automatically from textual radiology reports. We frame knowledge transfer as hyperparameter optimization, thus avoiding heuristic choices that are frequent in related works. We investigate the relationship between model size and TL, comparing a low-capacity VGG with a higher-capacity SEResNeXt. The task that we address is change detection in follow-up glioma imaging: we extracted 1693 T2-weighted magnetic resonance imaging difference maps from 183 patients, and classified them into stable or unstable according to tumor evolution. Weak labeling allowed us to increase dataset size more than 3-fold, and improve VGG classification results from 75% to 82% Area Under the ROC Curve (AUC) (p=0.04). Mixed training from scratch led to higher performance than fine-tuning or feature extraction. To assess generalizability, we also ran inference on an open dataset (BraTS-2015: 15 patients, 51 difference maps), reaching up to 76% AUC. Overall, results suggest that medical imaging problems may benefit from smaller models and different TL strategies with respect to computer vision problems, and that report-generated weak labels are effective in improving model performances. Code, in-house dataset and BraTS labels are released.
FaBiAN: A Fetal Brain magnetic resonance Acquisition Numerical phantom
Sep 06, 2021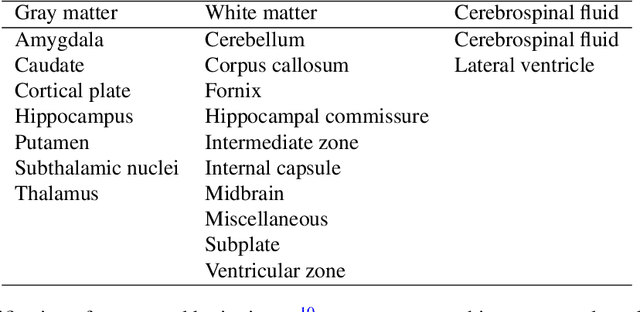
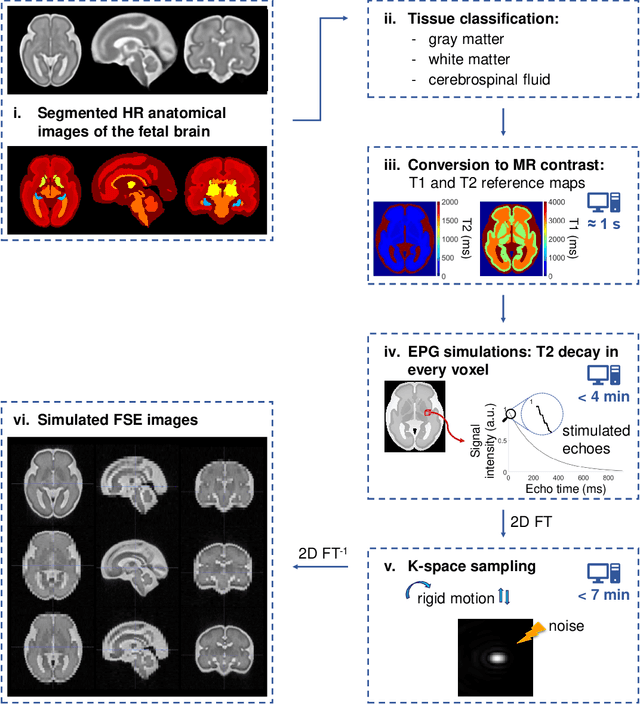

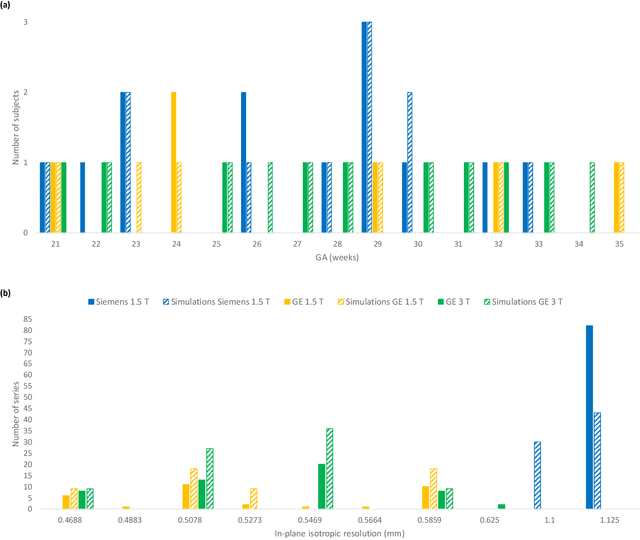
Abstract:Accurate characterization of in utero human brain maturation is critical as it involves complex and interconnected structural and functional processes that may influence health later in life. Magnetic resonance imaging is a powerful tool to investigate equivocal neurological patterns during fetal development. However, the number of acquisitions of satisfactory quality available in this cohort of sensitive subjects remains scarce, thus hindering the validation of advanced image processing techniques. Numerical phantoms can mitigate these limitations by providing a controlled environment with a known ground truth. In this work, we present FaBiAN, an open-source Fetal Brain magnetic resonance Acquisition Numerical phantom that simulates clinical T2-weighted fast spin echo sequences of the fetal brain. This unique tool is based on a general, flexible and realistic setup that includes stochastic fetal movements, thus providing images of the fetal brain throughout maturation comparable to clinical acquisitions. We demonstrate its value to evaluate the robustness and optimize the accuracy of an algorithm for super-resolution fetal brain magnetic resonance imaging from simulated motion-corrupted 2D low-resolution series as compared to a synthetic high-resolution reference volume. We also show that the images generated can complement clinical datasets to support data-intensive deep learning methods for fetal brain tissue segmentation.
Weak labels and anatomical knowledge: making deep learning practical for intracranial aneurysm detection in TOF-MRA
Mar 10, 2021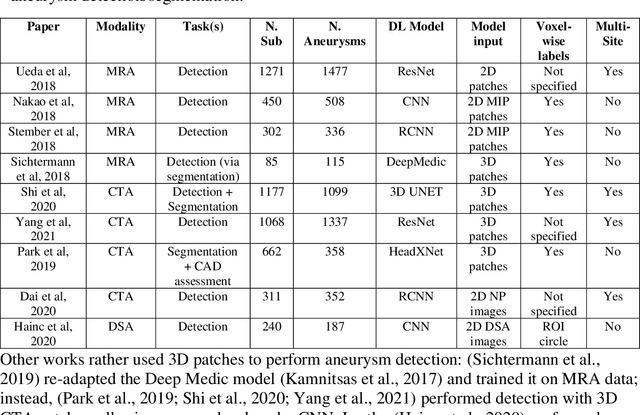
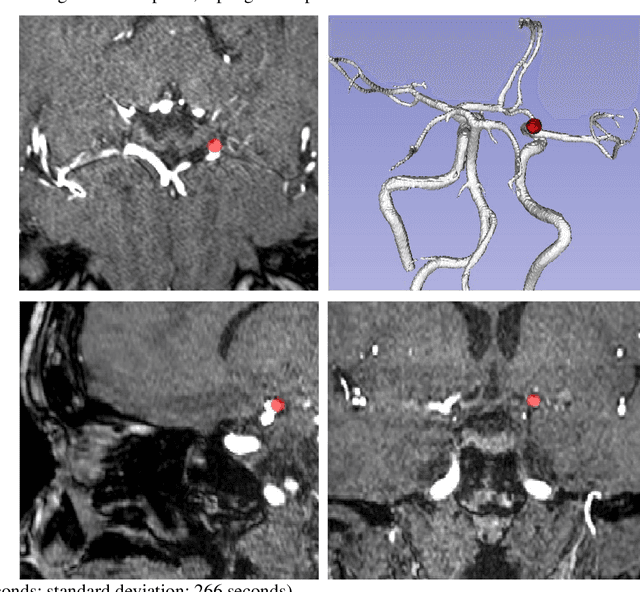
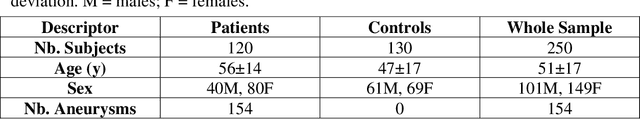
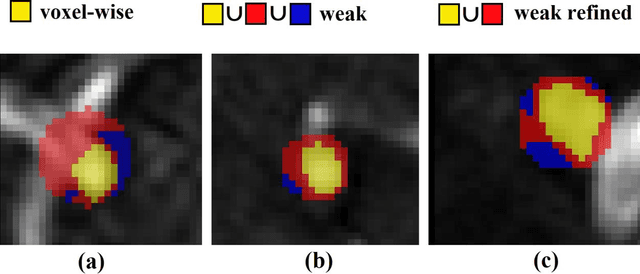
Abstract:Supervised segmentation algorithms yield state-of-the-art results for automated anomaly detection. However, these models require voxel-wise labels which are time-consuming to draw for medical experts. An interesting alternative to voxel-wise annotations is the use of weak labels: these can be coarse or oversized annotations that are less precise, but considerably faster to create. In this work, we address the task of brain aneurysm detection by developing a fully automated, deep neural network that is trained utilizing oversized weak labels. Furthermore, since aneurysms mainly occur in specific anatomical locations, we build our model leveraging the underlying anatomy of the brain vasculature both during training and inference. We apply our model to 250 subjects (120 patients, 130 controls) who underwent Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) and presented a total of 154 aneurysms. To assess the robustness of the algorithm, we participated in a MICCAI challenge for TOF-MRA data (93 patients, 20 controls, 125 aneurysms) which allowed us to obtain results also for subjects coming from a different institution. Our network achieves an average sensitivity of 77% on our in-house data, with a mean False Positive (FP) rate of 0.72 per patient. Instead, on the challenge data, we attain a sensitivity of 59% with a mean FP rate of 1.18, ranking in 7th/14 position for detection and in 4th/11 for segmentation on the open leaderboard. When computing detection performances with respect to aneurysms' risk of rupture, we found no statistical difference between two risk groups (p = 0.12), although the sensitivity for dangerous aneurysms was higher (78%). Our approach suggests that clinically useful sensitivity can be achieved using weak labels and exploiting prior anatomical knowledge; this expands the feasibility of deep learning studies to hospitals that have limited time and data.
An anatomically-informed 3D CNN for brain aneurysm classification with weak labels
Nov 27, 2020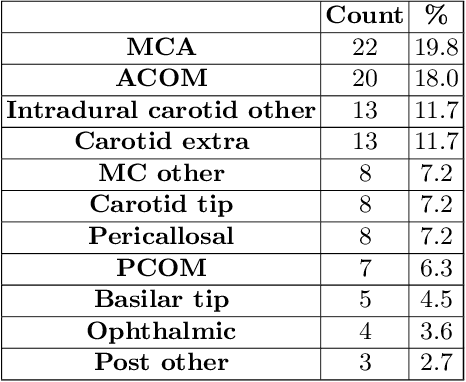
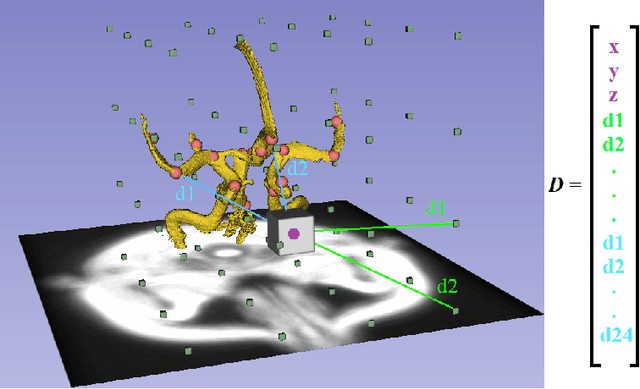
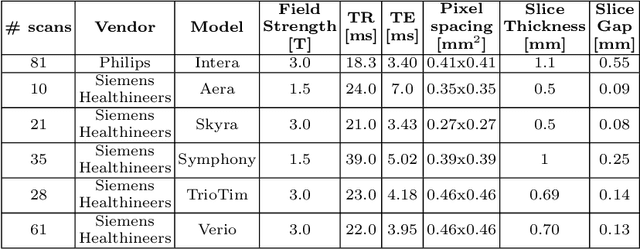
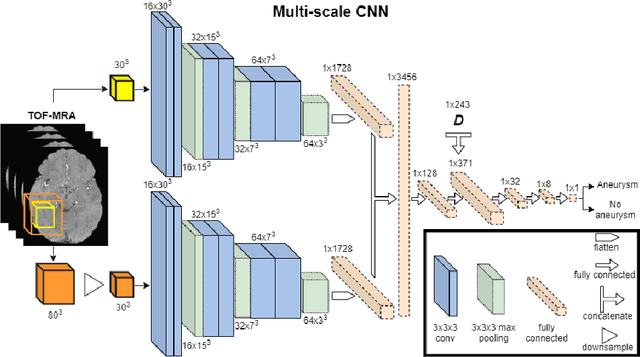
Abstract:A commonly adopted approach to carry out detection tasks in medical imaging is to rely on an initial segmentation. However, this approach strongly depends on voxel-wise annotations which are repetitive and time-consuming to draw for medical experts. An interesting alternative to voxel-wise masks are so-called "weak" labels: these can either be coarse or oversized annotations that are less precise, but noticeably faster to create. In this work, we address the task of brain aneurysm detection as a patch-wise binary classification with weak labels, in contrast to related studies that rather use supervised segmentation methods and voxel-wise delineations. Our approach comes with the non-trivial challenge of the data set creation: as for most focal diseases, anomalous patches (with aneurysm) are outnumbered by those showing no anomaly, and the two classes usually have different spatial distributions. To tackle this frequent scenario of inherently imbalanced, spatially skewed data sets, we propose a novel, anatomically-driven approach by using a multi-scale and multi-input 3D Convolutional Neural Network (CNN). We apply our model to 214 subjects (83 patients, 131 controls) who underwent Time-Of-Flight Magnetic Resonance Angiography (TOF-MRA) and presented a total of 111 unruptured cerebral aneurysms. We compare two strategies for negative patch sampling that have an increasing level of difficulty for the network and we show how this choice can strongly affect the results. To assess whether the added spatial information helps improving performances, we compare our anatomically-informed CNN with a baseline, spatially-agnostic CNN. When considering the more realistic and challenging scenario including vessel-like negative patches, the former model attains the highest classification results (accuracy$\simeq$95\%, AUROC$\simeq$0.95, AUPR$\simeq$0.71), thus outperforming the baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge