James Y. Zou
Accelerating Transformers with Spectrum-Preserving Token Merging
May 25, 2024



Abstract:Increasing the throughput of the Transformer architecture, a foundational component used in numerous state-of-the-art models for vision and language tasks (e.g., GPT, LLaVa), is an important problem in machine learning. One recent and effective strategy is to merge token representations within Transformer models, aiming to reduce computational and memory requirements while maintaining accuracy. Prior works have proposed algorithms based on Bipartite Soft Matching (BSM), which divides tokens into distinct sets and merges the top k similar tokens. However, these methods have significant drawbacks, such as sensitivity to token-splitting strategies and damage to informative tokens in later layers. This paper presents a novel paradigm called PiToMe, which prioritizes the preservation of informative tokens using an additional metric termed the energy score. This score identifies large clusters of similar tokens as high-energy, indicating potential candidates for merging, while smaller (unique and isolated) clusters are considered as low-energy and preserved. Experimental findings demonstrate that PiToMe saved from 40-60\% FLOPs of the base models while exhibiting superior off-the-shelf performance on image classification (0.5\% average performance drop of ViT-MAE-H compared to 2.6\% as baselines), image-text retrieval (0.3\% average performance drop of CLIP on Flickr30k compared to 4.5\% as others), and analogously in visual questions answering with LLaVa-7B. Furthermore, PiToMe is theoretically shown to preserve intrinsic spectral properties of the original token space under mild conditions
Mapping the Increasing Use of LLMs in Scientific Papers
Apr 01, 2024



Abstract:Scientific publishing lays the foundation of science by disseminating research findings, fostering collaboration, encouraging reproducibility, and ensuring that scientific knowledge is accessible, verifiable, and built upon over time. Recently, there has been immense speculation about how many people are using large language models (LLMs) like ChatGPT in their academic writing, and to what extent this tool might have an effect on global scientific practices. However, we lack a precise measure of the proportion of academic writing substantially modified or produced by LLMs. To address this gap, we conduct the first systematic, large-scale analysis across 950,965 papers published between January 2020 and February 2024 on the arXiv, bioRxiv, and Nature portfolio journals, using a population-level statistical framework to measure the prevalence of LLM-modified content over time. Our statistical estimation operates on the corpus level and is more robust than inference on individual instances. Our findings reveal a steady increase in LLM usage, with the largest and fastest growth observed in Computer Science papers (up to 17.5%). In comparison, Mathematics papers and the Nature portfolio showed the least LLM modification (up to 6.3%). Moreover, at an aggregate level, our analysis reveals that higher levels of LLM-modification are associated with papers whose first authors post preprints more frequently, papers in more crowded research areas, and papers of shorter lengths. Our findings suggests that LLMs are being broadly used in scientific writings.
Monitoring AI-Modified Content at Scale: A Case Study on the Impact of ChatGPT on AI Conference Peer Reviews
Mar 11, 2024



Abstract:We present an approach for estimating the fraction of text in a large corpus which is likely to be substantially modified or produced by a large language model (LLM). Our maximum likelihood model leverages expert-written and AI-generated reference texts to accurately and efficiently examine real-world LLM-use at the corpus level. We apply this approach to a case study of scientific peer review in AI conferences that took place after the release of ChatGPT: ICLR 2024, NeurIPS 2023, CoRL 2023 and EMNLP 2023. Our results suggest that between 6.5% and 16.9% of text submitted as peer reviews to these conferences could have been substantially modified by LLMs, i.e. beyond spell-checking or minor writing updates. The circumstances in which generated text occurs offer insight into user behavior: the estimated fraction of LLM-generated text is higher in reviews which report lower confidence, were submitted close to the deadline, and from reviewers who are less likely to respond to author rebuttals. We also observe corpus-level trends in generated text which may be too subtle to detect at the individual level, and discuss the implications of such trends on peer review. We call for future interdisciplinary work to examine how LLM use is changing our information and knowledge practices.
Protein structure generation via folding diffusion
Sep 30, 2022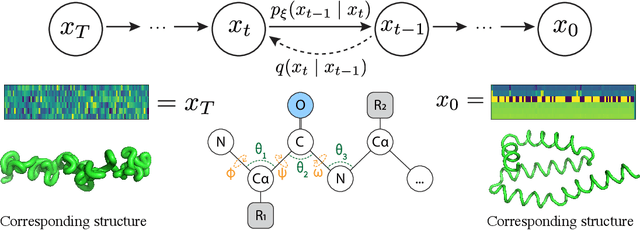
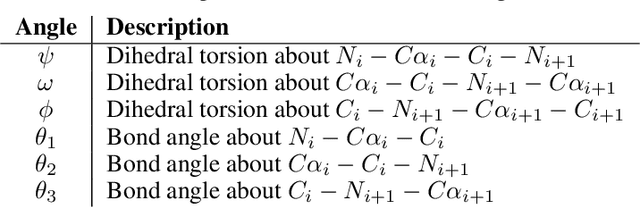
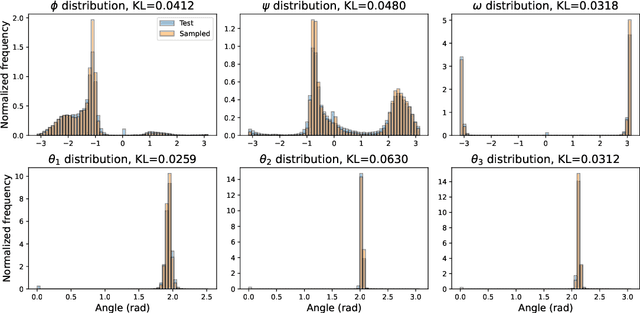
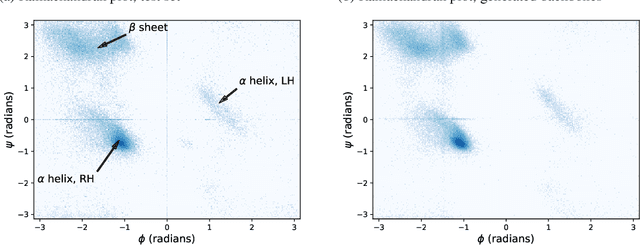
Abstract:The ability to computationally generate novel yet physically foldable protein structures could lead to new biological discoveries and new treatments targeting yet incurable diseases. Despite recent advances in protein structure prediction, directly generating diverse, novel protein structures from neural networks remains difficult. In this work, we present a new diffusion-based generative model that designs protein backbone structures via a procedure that mirrors the native folding process. We describe protein backbone structure as a series of consecutive angles capturing the relative orientation of the constituent amino acid residues, and generate new structures by denoising from a random, unfolded state towards a stable folded structure. Not only does this mirror how proteins biologically twist into energetically favorable conformations, the inherent shift and rotational invariance of this representation crucially alleviates the need for complex equivariant networks. We train a denoising diffusion probabilistic model with a simple transformer backbone and demonstrate that our resulting model unconditionally generates highly realistic protein structures with complexity and structural patterns akin to those of naturally-occurring proteins. As a useful resource, we release the first open-source codebase and trained models for protein structure diffusion.
High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy with Cardiovascular Deep Learning
Jun 23, 2021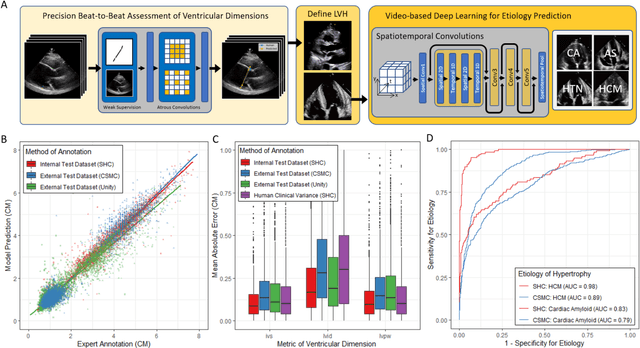
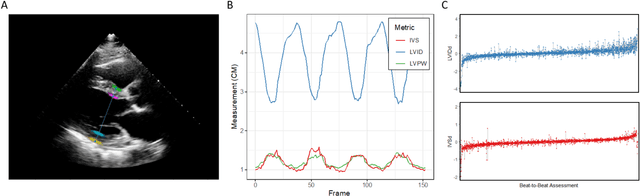
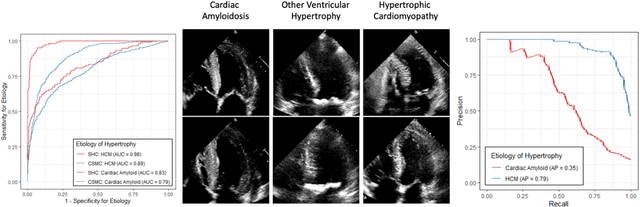
Abstract:Left ventricular hypertrophy (LVH) results from chronic remodeling caused by a broad range of systemic and cardiovascular disease including hypertension, aortic stenosis, hypertrophic cardiomyopathy, and cardiac amyloidosis. Early detection and characterization of LVH can significantly impact patient care but is limited by under-recognition of hypertrophy, measurement error and variability, and difficulty differentiating etiologies of LVH. To overcome this challenge, we present EchoNet-LVH - a deep learning workflow that automatically quantifies ventricular hypertrophy with precision equal to human experts and predicts etiology of LVH. Trained on 28,201 echocardiogram videos, our model accurately measures intraventricular wall thickness (mean absolute error [MAE] 1.4mm, 95% CI 1.2-1.5mm), left ventricular diameter (MAE 2.4mm, 95% CI 2.2-2.6mm), and posterior wall thickness (MAE 1.2mm, 95% CI 1.1-1.3mm) and classifies cardiac amyloidosis (area under the curve of 0.83) and hypertrophic cardiomyopathy (AUC 0.98) from other etiologies of LVH. In external datasets from independent domestic and international healthcare systems, EchoNet-LVH accurately quantified ventricular parameters (R2 of 0.96 and 0.90 respectively) and detected cardiac amyloidosis (AUC 0.79) and hypertrophic cardiomyopathy (AUC 0.89) on the domestic external validation site. Leveraging measurements across multiple heart beats, our model can more accurately identify subtle changes in LV geometry and its causal etiologies. Compared to human experts, EchoNet-LVH is fully automated, allowing for reproducible, precise measurements, and lays the foundation for precision diagnosis of cardiac hypertrophy. As a resource to promote further innovation, we also make publicly available a large dataset of 23,212 annotated echocardiogram videos.
Crowdsourcing Feature Discovery via Adaptively Chosen Comparisons
Mar 31, 2015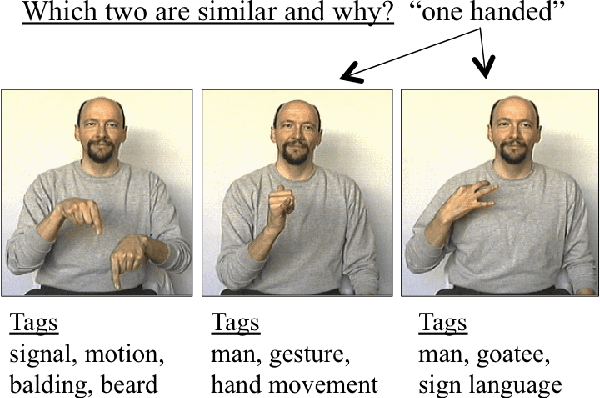

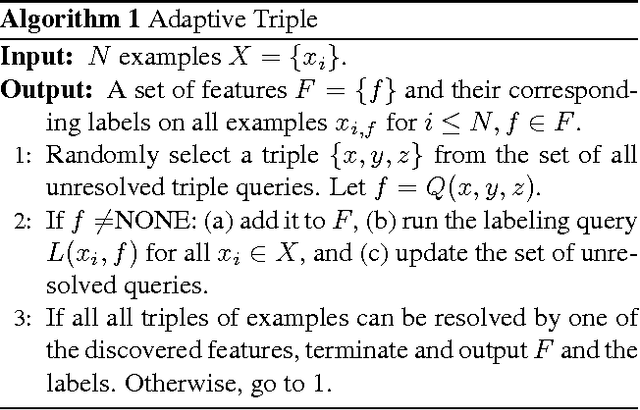

Abstract:We introduce an unsupervised approach to efficiently discover the underlying features in a data set via crowdsourcing. Our queries ask crowd members to articulate a feature common to two out of three displayed examples. In addition we also ask the crowd to provide binary labels to the remaining examples based on the discovered features. The triples are chosen adaptively based on the labels of the previously discovered features on the data set. In two natural models of features, hierarchical and independent, we show that a simple adaptive algorithm, using "two-out-of-three" similarity queries, recovers all features with less labor than any nonadaptive algorithm. Experimental results validate the theoretical findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge