Alex X. Lu
Causal integration of chemical structures improves representations of microscopy images for morphological profiling
Apr 13, 2025Abstract:Recent advances in self-supervised deep learning have improved our ability to quantify cellular morphological changes in high-throughput microscopy screens, a process known as morphological profiling. However, most current methods only learn from images, despite many screens being inherently multimodal, as they involve both a chemical or genetic perturbation as well as an image-based readout. We hypothesized that incorporating chemical compound structure during self-supervised pre-training could improve learned representations of images in high-throughput microscopy screens. We introduce a representation learning framework, MICON (Molecular-Image Contrastive Learning), that models chemical compounds as treatments that induce counterfactual transformations of cell phenotypes. MICON significantly outperforms classical hand-crafted features such as CellProfiler and existing deep-learning-based representation learning methods in challenging evaluation settings where models must identify reproducible effects of drugs across independent replicates and data-generating centers. We demonstrate that incorporating chemical compound information into the learning process provides consistent improvements in our evaluation setting and that modeling compounds specifically as treatments in a causal framework outperforms approaches that directly align images and compounds in a single representation space. Our findings point to a new direction for representation learning in morphological profiling, suggesting that methods should explicitly account for the multimodal nature of microscopy screening data.
ASL STEM Wiki: Dataset and Benchmark for Interpreting STEM Articles
Nov 08, 2024Abstract:Deaf and hard-of-hearing (DHH) students face significant barriers in accessing science, technology, engineering, and mathematics (STEM) education, notably due to the scarcity of STEM resources in signed languages. To help address this, we introduce ASL STEM Wiki: a parallel corpus of 254 Wikipedia articles on STEM topics in English, interpreted into over 300 hours of American Sign Language (ASL). ASL STEM Wiki is the first continuous signing dataset focused on STEM, facilitating the development of AI resources for STEM education in ASL. We identify several use cases of ASL STEM Wiki with human-centered applications. For example, because this dataset highlights the frequent use of fingerspelling for technical concepts, which inhibits DHH students' ability to learn, we develop models to identify fingerspelled words -- which can later be used to query for appropriate ASL signs to suggest to interpreters.
Systemic Biases in Sign Language AI Research: A Deaf-Led Call to Reevaluate Research Agendas
Mar 05, 2024Abstract:Growing research in sign language recognition, generation, and translation AI has been accompanied by calls for ethical development of such technologies. While these works are crucial to helping individual researchers do better, there is a notable lack of discussion of systemic biases or analysis of rhetoric that shape the research questions and methods in the field, especially as it remains dominated by hearing non-signing researchers. Therefore, we conduct a systematic review of 101 recent papers in sign language AI. Our analysis identifies significant biases in the current state of sign language AI research, including an overfocus on addressing perceived communication barriers, a lack of use of representative datasets, use of annotations lacking linguistic foundations, and development of methods that build on flawed models. We take the position that the field lacks meaningful input from Deaf stakeholders, and is instead driven by what decisions are the most convenient or perceived as important to hearing researchers. We end with a call to action: the field must make space for Deaf researchers to lead the conversation in sign language AI.
ASL Citizen: A Community-Sourced Dataset for Advancing Isolated Sign Language Recognition
Apr 12, 2023



Abstract:Sign languages are used as a primary language by approximately 70 million D/deaf people world-wide. However, most communication technologies operate in spoken and written languages, creating inequities in access. To help tackle this problem, we release ASL Citizen, the largest Isolated Sign Language Recognition (ISLR) dataset to date, collected with consent and containing 83,912 videos for 2,731 distinct signs filmed by 52 signers in a variety of environments. We propose that this dataset be used for sign language dictionary retrieval for American Sign Language (ASL), where a user demonstrates a sign to their own webcam with the aim of retrieving matching signs from a dictionary. We show that training supervised machine learning classifiers with our dataset greatly advances the state-of-the-art on metrics relevant for dictionary retrieval, achieving, for instance, 62% accuracy and a recall-at-10 of 90%, evaluated entirely on videos of users who are not present in the training or validation sets. An accessible PDF of this article is available at https://aashakadesai.github.io/research/ASL_Dataset__arxiv_.pdf
Protein structure generation via folding diffusion
Sep 30, 2022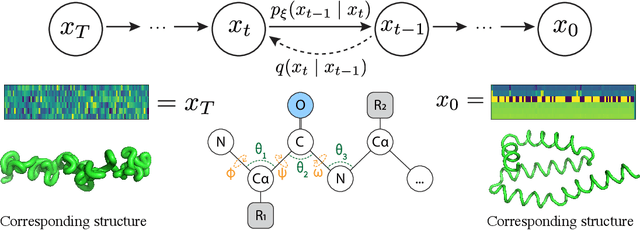
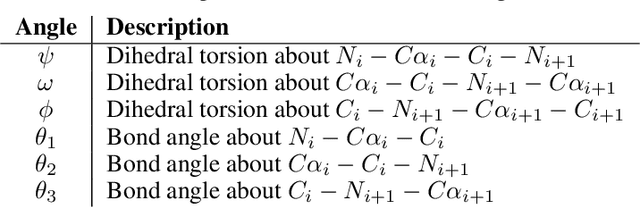
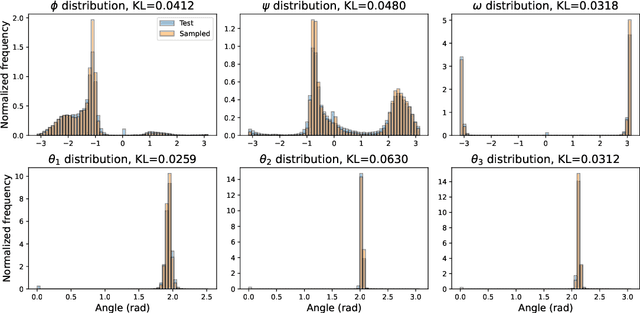
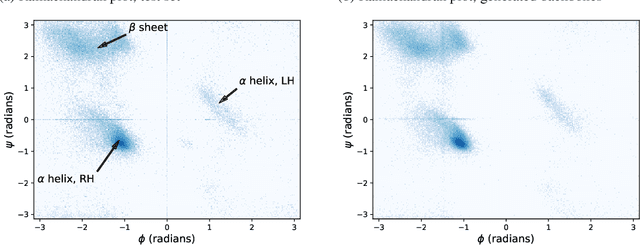
Abstract:The ability to computationally generate novel yet physically foldable protein structures could lead to new biological discoveries and new treatments targeting yet incurable diseases. Despite recent advances in protein structure prediction, directly generating diverse, novel protein structures from neural networks remains difficult. In this work, we present a new diffusion-based generative model that designs protein backbone structures via a procedure that mirrors the native folding process. We describe protein backbone structure as a series of consecutive angles capturing the relative orientation of the constituent amino acid residues, and generate new structures by denoising from a random, unfolded state towards a stable folded structure. Not only does this mirror how proteins biologically twist into energetically favorable conformations, the inherent shift and rotational invariance of this representation crucially alleviates the need for complex equivariant networks. We train a denoising diffusion probabilistic model with a simple transformer backbone and demonstrate that our resulting model unconditionally generates highly realistic protein structures with complexity and structural patterns akin to those of naturally-occurring proteins. As a useful resource, we release the first open-source codebase and trained models for protein structure diffusion.
CytoImageNet: A large-scale pretraining dataset for bioimage transfer learning
Nov 24, 2021
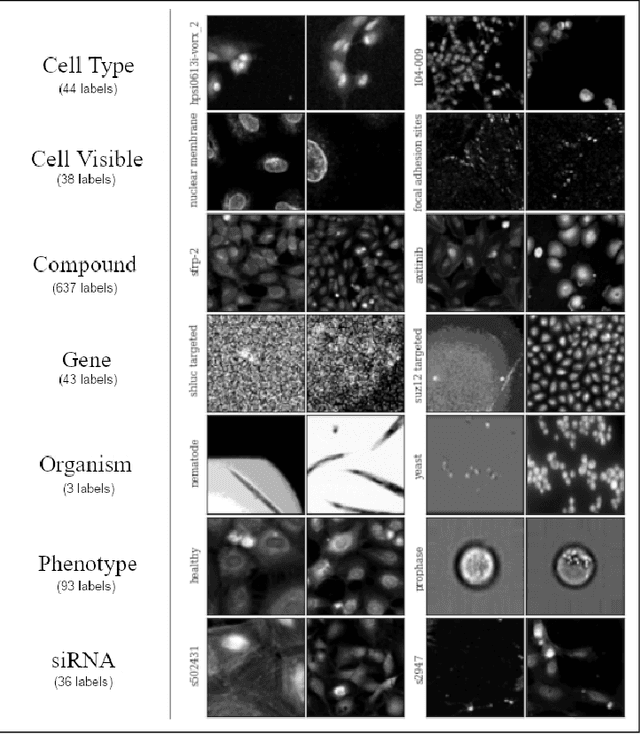
Abstract:Motivation: In recent years, image-based biological assays have steadily become high-throughput, sparking a need for fast automated methods to extract biologically-meaningful information from hundreds of thousands of images. Taking inspiration from the success of ImageNet, we curate CytoImageNet, a large-scale dataset of openly-sourced and weakly-labeled microscopy images (890K images, 894 classes). Pretraining on CytoImageNet yields features that are competitive to ImageNet features on downstream microscopy classification tasks. We show evidence that CytoImageNet features capture information not available in ImageNet-trained features. The dataset is made available at https://www.kaggle.com/stanleyhua/cytoimagenet.
Random Embeddings and Linear Regression can Predict Protein Function
Apr 25, 2021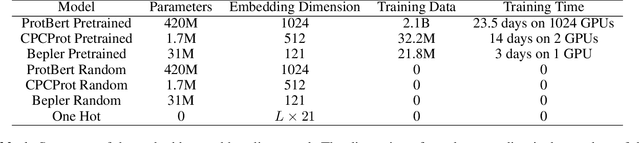
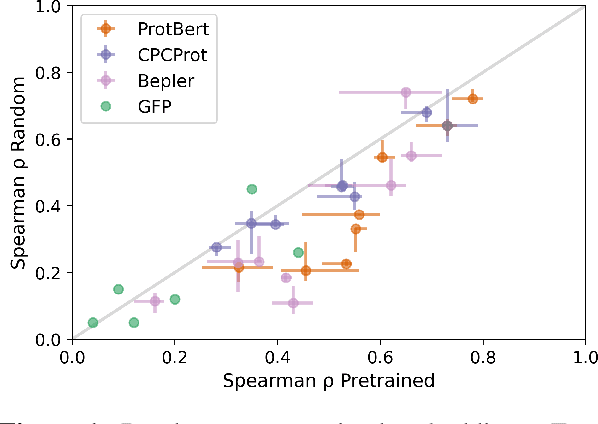
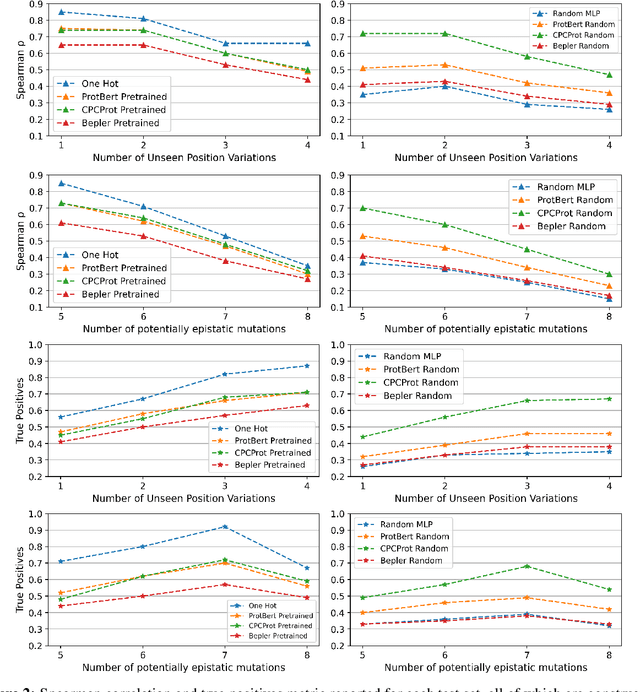
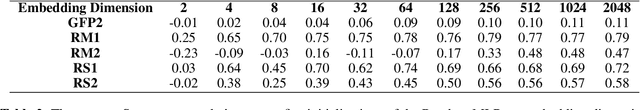
Abstract:Large self-supervised models pretrained on millions of protein sequences have recently gained popularity in generating embeddings of protein sequences for protein function prediction. However, the absence of random baselines makes it difficult to conclude whether pretraining has learned useful information for protein function prediction. Here we show that one-hot encoding and random embeddings, both of which do not require any pretraining, are strong baselines for protein function prediction across 14 diverse sequence-to-function tasks.
Evolution Is All You Need: Phylogenetic Augmentation for Contrastive Learning
Dec 25, 2020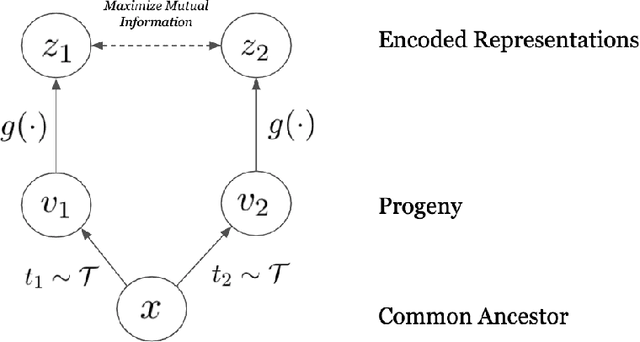
Abstract:Self-supervised representation learning of biological sequence embeddings alleviates computational resource constraints on downstream tasks while circumventing expensive experimental label acquisition. However, existing methods mostly borrow directly from large language models designed for NLP, rather than with bioinformatics philosophies in mind. Recently, contrastive mutual information maximization methods have achieved state-of-the-art representations for ImageNet. In this perspective piece, we discuss how viewing evolution as natural sequence augmentation and maximizing information across phylogenetic "noisy channels" is a biologically and theoretically desirable objective for pretraining encoders. We first provide a review of current contrastive learning literature, then provide an illustrative example where we show that contrastive learning using evolutionary augmentation can be used as a representation learning objective which maximizes the mutual information between biological sequences and their conserved function, and finally outline rationale for this approach.
The Cells Out of Sample dataset and benchmarks for measuring out-of-sample generalization of image classifiers
Jun 17, 2019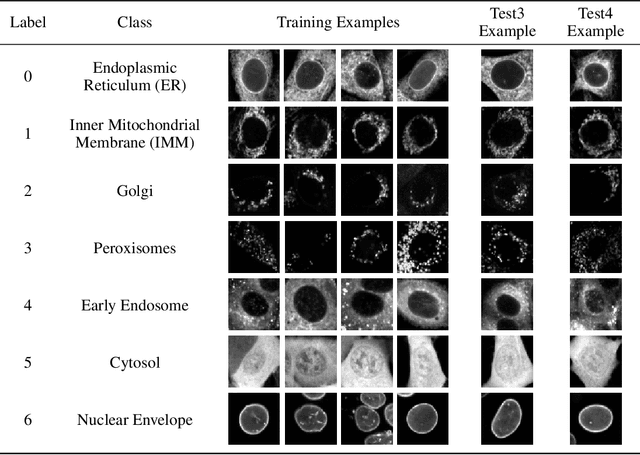
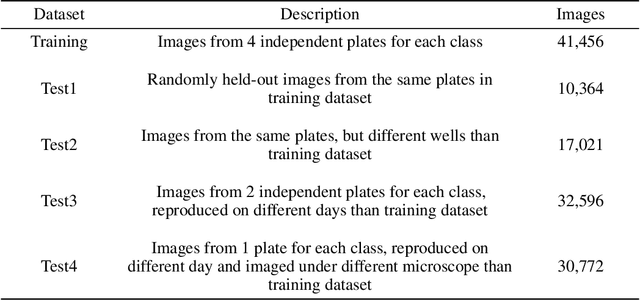
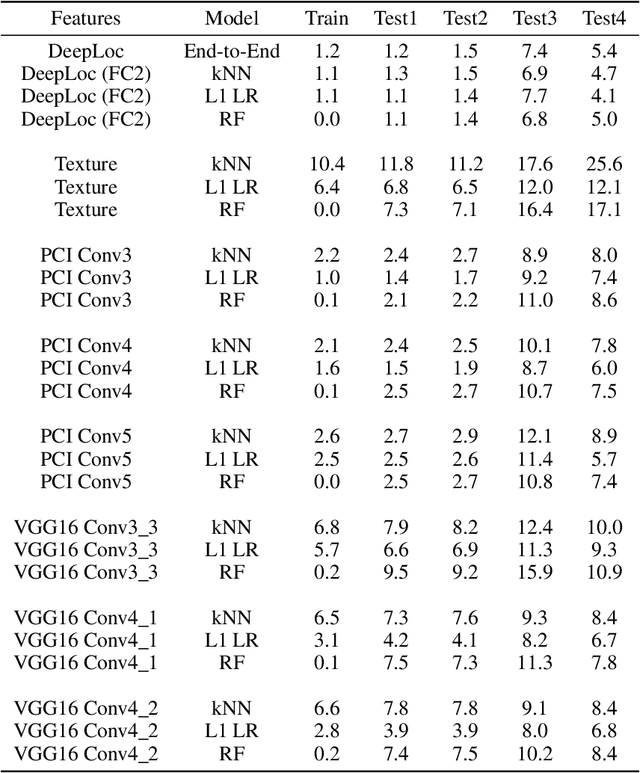
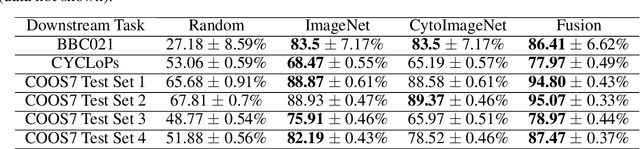
Abstract:Understanding if classifiers generalize to out-of-sample datasets is a central problem in machine learning. Microscopy images provide a standardized way to measure the generalization capacity of image classifiers, as we can image the same classes of objects under increasingly divergent, but controlled factors of variation. We created a public dataset of 132,209 images of mouse cells, COOS-7 (Cells Out Of Sample 7-Class). COOS-7 provides a classification setting where four test datasets have increasing degrees of covariate shift: some images are random subsets of the training data, while others are from experiments reproduced months later and imaged by different instruments. We benchmarked a range of classification models using different representations, including transferred neural network features, end-to-end classification with a supervised deep CNN, and features from a self-supervised CNN. While most classifiers perform well on test datasets similar to the training dataset, all classifiers failed to generalize their performance to datasets with greater covariate shifts. These baselines highlight the challenges of covariate shifts in image data, and establish metrics for improving the generalization capacity of image classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge