J. Alison Noble
Image quality assessment for machine learning tasks using meta-reinforcement learning
Mar 27, 2022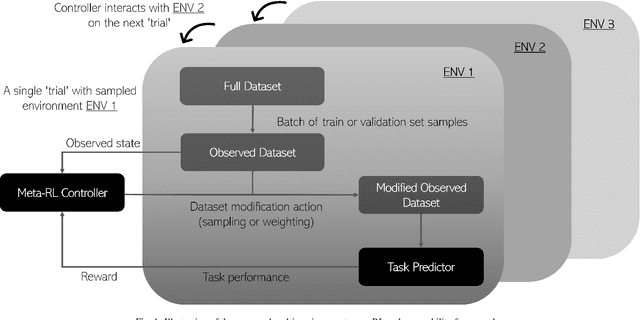
Abstract:In this paper, we consider image quality assessment (IQA) as a measure of how images are amenable with respect to a given downstream task, or task amenability. When the task is performed using machine learning algorithms, such as a neural-network-based task predictor for image classification or segmentation, the performance of the task predictor provides an objective estimate of task amenability. In this work, we use an IQA controller to predict the task amenability which, itself being parameterised by neural networks, can be trained simultaneously with the task predictor. We further develop a meta-reinforcement learning framework to improve the adaptability for both IQA controllers and task predictors, such that they can be fine-tuned efficiently on new datasets or meta-tasks. We demonstrate the efficacy of the proposed task-specific, adaptable IQA approach, using two clinical applications for ultrasound-guided prostate intervention and pneumonia detection on X-ray images.
* Accepted to Medical Image Analysis; Final published version available at: https://doi.org/10.1016/j.media.2022.102427
Facial Anatomical Landmark Detection using Regularized Transfer Learning with Application to Fetal Alcohol Syndrome Recognition
Sep 12, 2021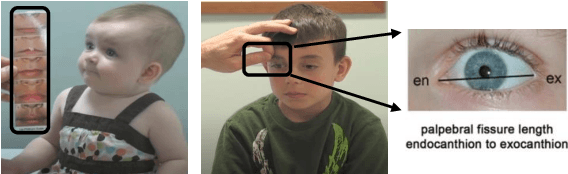
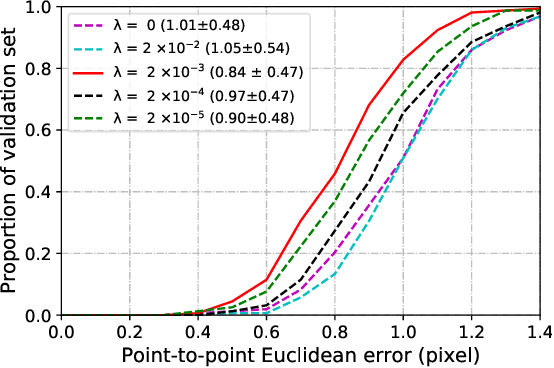
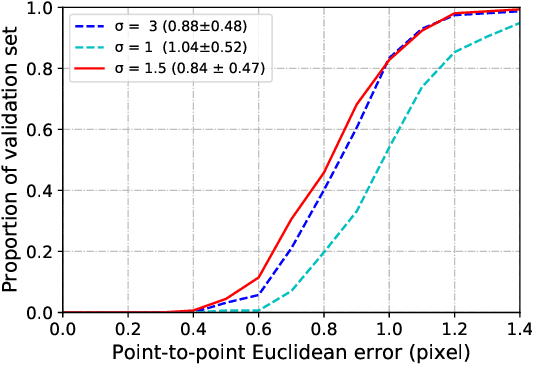
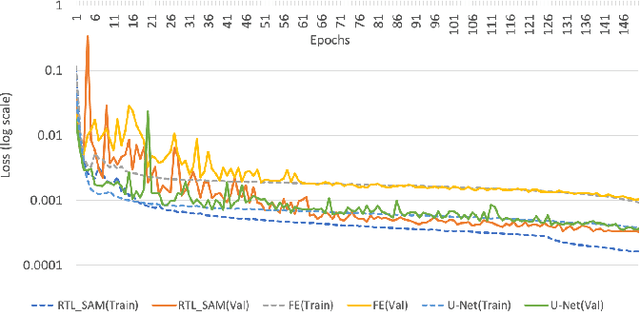
Abstract:Fetal alcohol syndrome (FAS) caused by prenatal alcohol exposure can result in a series of cranio-facial anomalies, and behavioral and neurocognitive problems. Current diagnosis of FAS is typically done by identifying a set of facial characteristics, which are often obtained by manual examination. Anatomical landmark detection, which provides rich geometric information, is important to detect the presence of FAS associated facial anomalies. This imaging application is characterized by large variations in data appearance and limited availability of labeled data. Current deep learning-based heatmap regression methods designed for facial landmark detection in natural images assume availability of large datasets and are therefore not wellsuited for this application. To address this restriction, we develop a new regularized transfer learning approach that exploits the knowledge of a network learned on large facial recognition datasets. In contrast to standard transfer learning which focuses on adjusting the pre-trained weights, the proposed learning approach regularizes the model behavior. It explicitly reuses the rich visual semantics of a domain-similar source model on the target task data as an additional supervisory signal for regularizing landmark detection optimization. Specifically, we develop four regularization constraints for the proposed transfer learning, including constraining the feature outputs from classification and intermediate layers, as well as matching activation attention maps in both spatial and channel levels. Experimental evaluation on a collected clinical imaging dataset demonstrate that the proposed approach can effectively improve model generalizability under limited training samples, and is advantageous to other approaches in the literature.
Adaptable image quality assessment using meta-reinforcement learning of task amenability
Jul 31, 2021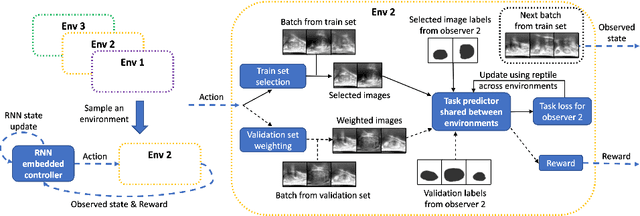
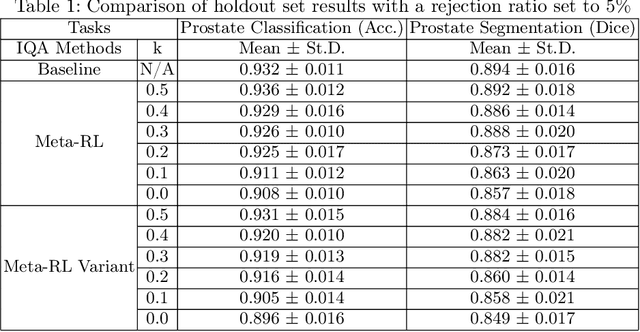
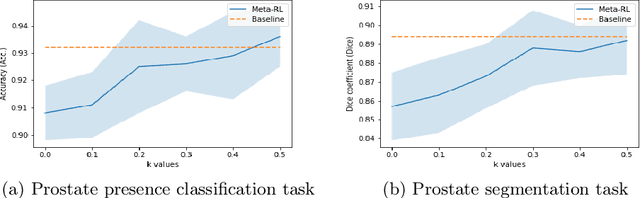
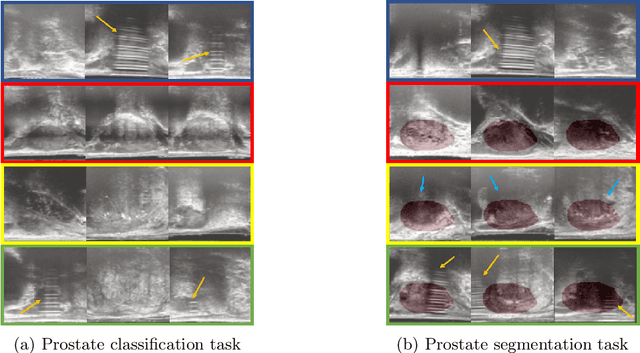
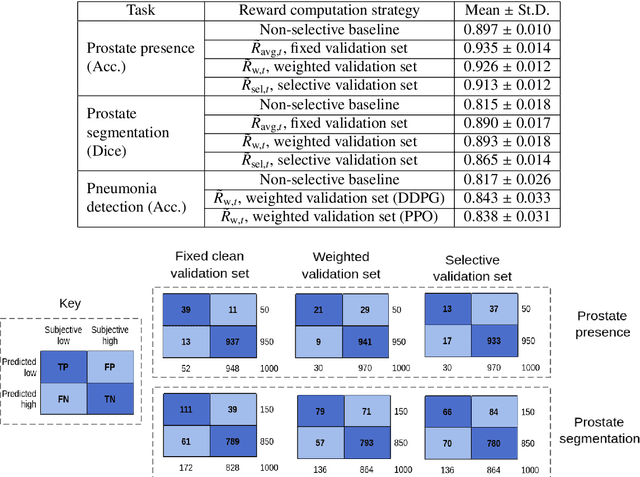
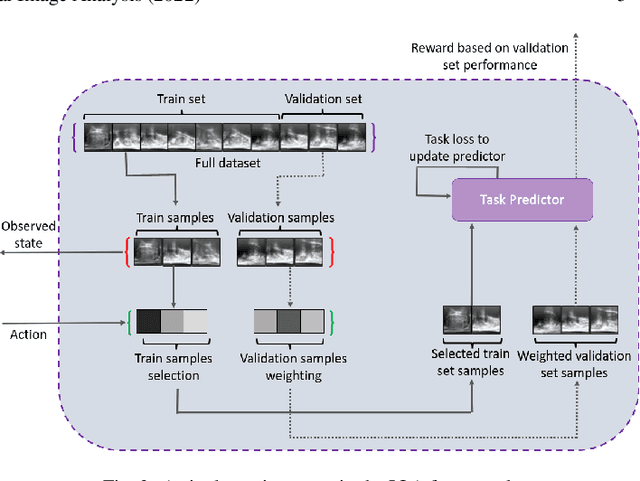
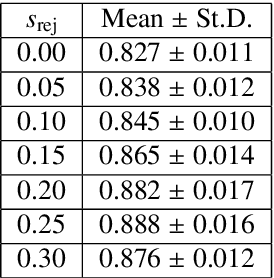
Abstract:The performance of many medical image analysis tasks are strongly associated with image data quality. When developing modern deep learning algorithms, rather than relying on subjective (human-based) image quality assessment (IQA), task amenability potentially provides an objective measure of task-specific image quality. To predict task amenability, an IQA agent is trained using reinforcement learning (RL) with a simultaneously optimised task predictor, such as a classification or segmentation neural network. In this work, we develop transfer learning or adaptation strategies to increase the adaptability of both the IQA agent and the task predictor so that they are less dependent on high-quality, expert-labelled training data. The proposed transfer learning strategy re-formulates the original RL problem for task amenability in a meta-reinforcement learning (meta-RL) framework. The resulting algorithm facilitates efficient adaptation of the agent to different definitions of image quality, each with its own Markov decision process environment including different images, labels and an adaptable task predictor. Our work demonstrates that the IQA agents pre-trained on non-expert task labels can be adapted to predict task amenability as defined by expert task labels, using only a small set of expert labels. Using 6644 clinical ultrasound images from 249 prostate cancer patients, our results for image classification and segmentation tasks show that the proposed IQA method can be adapted using data with as few as respective 19.7% and 29.6% expert-reviewed consensus labels and still achieve comparable IQA and task performance, which would otherwise require a training dataset with 100% expert labels.
Principled Ultrasound Data Augmentation for Classification of Standard Planes
Mar 14, 2021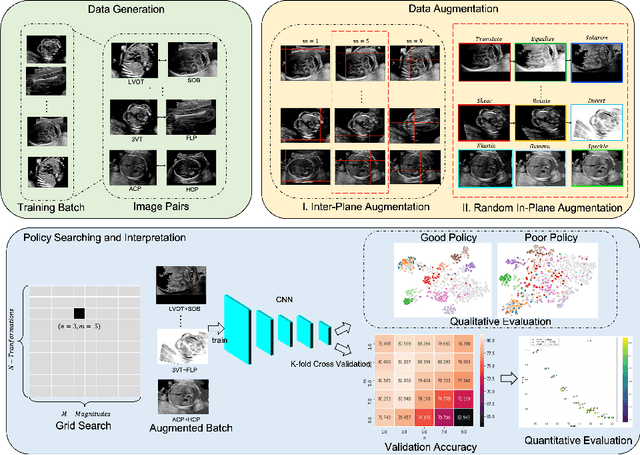
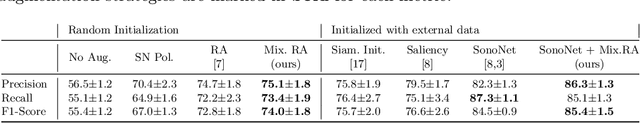
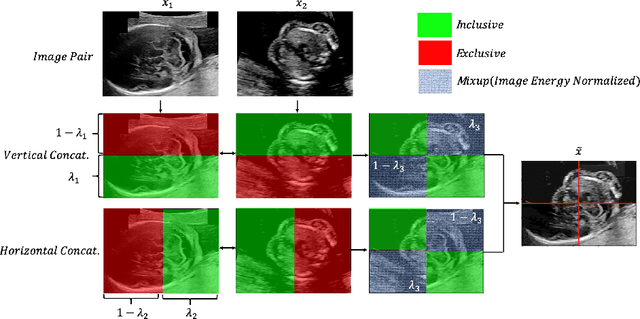
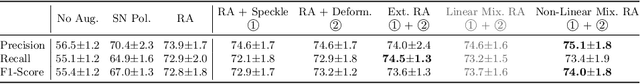
Abstract:Deep learning models with large learning capacities often overfit to medical imaging datasets. This is because training sets are often relatively small due to the significant time and financial costs incurred in medical data acquisition and labelling. Data augmentation is therefore often used to expand the availability of training data and to increase generalization. However, augmentation strategies are often chosen on an ad-hoc basis without justification. In this paper, we present an augmentation policy search method with the goal of improving model classification performance. We include in the augmentation policy search additional transformations that are often used in medical image analysis and evaluate their performance. In addition, we extend the augmentation policy search to include non-linear mixed-example data augmentation strategies. Using these learned policies, we show that principled data augmentation for medical image model training can lead to significant improvements in ultrasound standard plane detection, with an an average F1-score improvement of 7.0% overall over naive data augmentation strategies in ultrasound fetal standard plane classification. We find that the learned representations of ultrasound images are better clustered and defined with optimized data augmentation.
Cross-Task Representation Learning for Anatomical Landmark Detection
Sep 28, 2020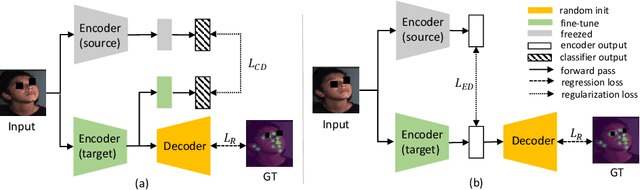
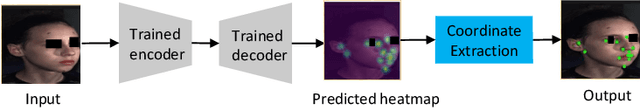
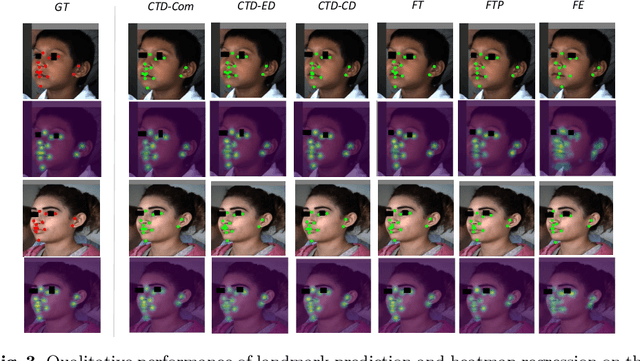
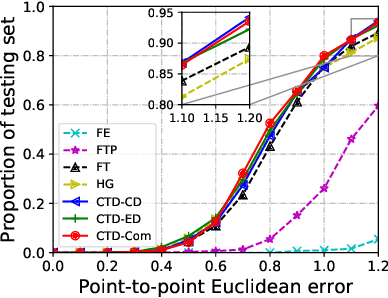
Abstract:Recently, there is an increasing demand for automatically detecting anatomical landmarks which provide rich structural information to facilitate subsequent medical image analysis. Current methods related to this task often leverage the power of deep neural networks, while a major challenge in fine tuning such models in medical applications arises from insufficient number of labeled samples. To address this, we propose to regularize the knowledge transfer across source and target tasks through cross-task representation learning. The proposed method is demonstrated for extracting facial anatomical landmarks which facilitate the diagnosis of fetal alcohol syndrome. The source and target tasks in this work are face recognition and landmark detection, respectively. The main idea of the proposed method is to retain the feature representations of the source model on the target task data, and to leverage them as an additional source of supervisory signals for regularizing the target model learning, thereby improving its performance under limited training samples. Concretely, we present two approaches for the proposed representation learning by constraining either final or intermediate model features on the target model. Experimental results on a clinical face image dataset demonstrate that the proposed approach works well with few labeled data, and outperforms other compared approaches.
Longitudinal Image Registration with Temporal-order and Subject-specificity Discrimination
Aug 29, 2020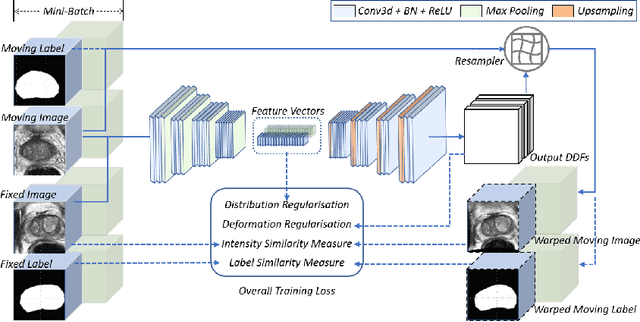

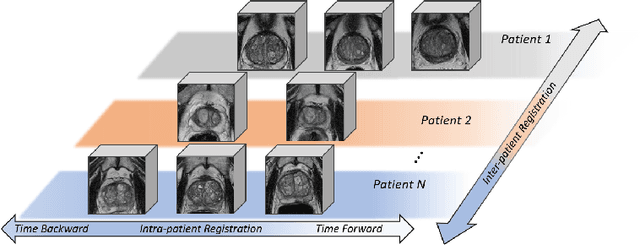
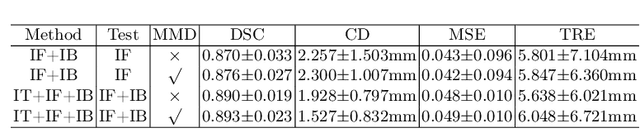
Abstract:Morphological analysis of longitudinal MR images plays a key role in monitoring disease progression for prostate cancer patients, who are placed under an active surveillance program. In this paper, we describe a learning-based image registration algorithm to quantify changes on regions of interest between a pair of images from the same patient, acquired at two different time points. Combining intensity-based similarity and gland segmentation as weak supervision, the population-data-trained registration networks significantly lowered the target registration errors (TREs) on holdout patient data, compared with those before registration and those from an iterative registration algorithm. Furthermore, this work provides a quantitative analysis on several longitudinal-data-sampling strategies and, in turn, we propose a novel regularisation method based on maximum mean discrepancy, between differently-sampled training image pairs. Based on 216 3D MR images from 86 patients, we report a mean TRE of 5.6 mm and show statistically significant differences between the different training data sampling strategies.
Self-Supervised Ultrasound to MRI Fetal Brain Image Synthesis
Aug 19, 2020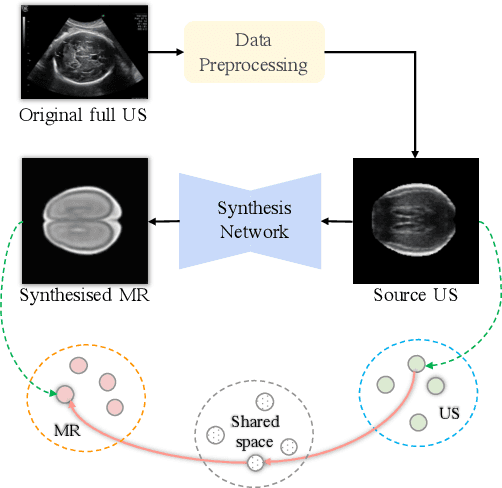
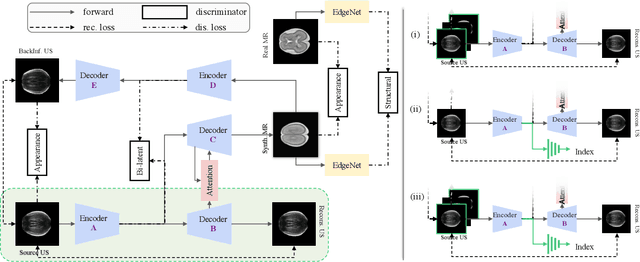
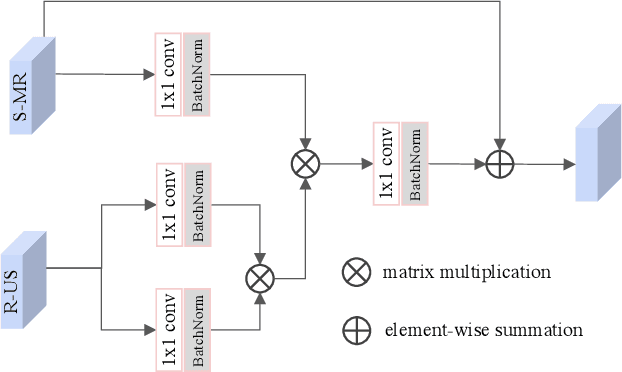
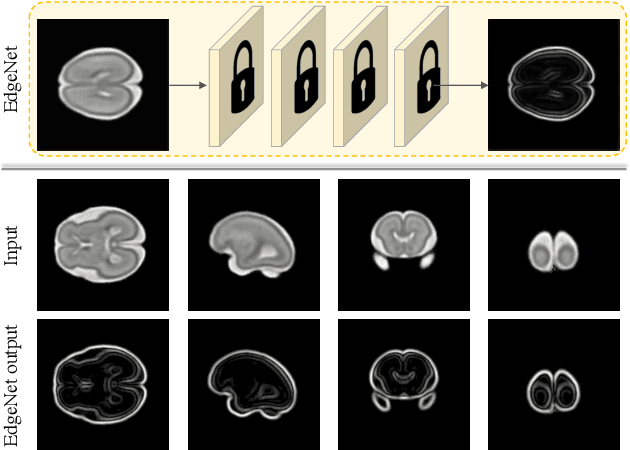
Abstract:Fetal brain magnetic resonance imaging (MRI) offers exquisite images of the developing brain but is not suitable for second-trimester anomaly screening, for which ultrasound (US) is employed. Although expert sonographers are adept at reading US images, MR images which closely resemble anatomical images are much easier for non-experts to interpret. Thus in this paper we propose to generate MR-like images directly from clinical US images. In medical image analysis such a capability is potentially useful as well, for instance for automatic US-MRI registration and fusion. The proposed model is end-to-end trainable and self-supervised without any external annotations. Specifically, based on an assumption that the US and MRI data share a similar anatomical latent space, we first utilise a network to extract the shared latent features, which are then used for MRI synthesis. Since paired data is unavailable for our study (and rare in practice), pixel-level constraints are infeasible to apply. We instead propose to enforce the distributions to be statistically indistinguishable, by adversarial learning in both the image domain and feature space. To regularise the anatomical structures between US and MRI during synthesis, we further propose an adversarial structural constraint. A new cross-modal attention technique is proposed to utilise non-local spatial information, by encouraging multi-modal knowledge fusion and propagation. We extend the approach to consider the case where 3D auxiliary information (e.g., 3D neighbours and a 3D location index) from volumetric data is also available, and show that this improves image synthesis. The proposed approach is evaluated quantitatively and qualitatively with comparison to real fetal MR images and other approaches to synthesis, demonstrating its feasibility of synthesising realistic MR images.
Self-supervised Contrastive Video-Speech Representation Learning for Ultrasound
Aug 14, 2020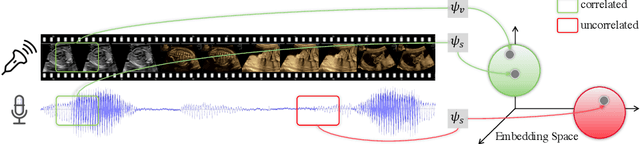


Abstract:In medical imaging, manual annotations can be expensive to acquire and sometimes infeasible to access, making conventional deep learning-based models difficult to scale. As a result, it would be beneficial if useful representations could be derived from raw data without the need for manual annotations. In this paper, we propose to address the problem of self-supervised representation learning with multi-modal ultrasound video-speech raw data. For this case, we assume that there is a high correlation between the ultrasound video and the corresponding narrative speech audio of the sonographer. In order to learn meaningful representations, the model needs to identify such correlation and at the same time understand the underlying anatomical features. We designed a framework to model the correspondence between video and audio without any kind of human annotations. Within this framework, we introduce cross-modal contrastive learning and an affinity-aware self-paced learning scheme to enhance correlation modelling. Experimental evaluations on multi-modal fetal ultrasound video and audio show that the proposed approach is able to learn strong representations and transfers well to downstream tasks of standard plane detection and eye-gaze prediction.
Automatic Probe Movement Guidance for Freehand Obstetric Ultrasound
Jul 08, 2020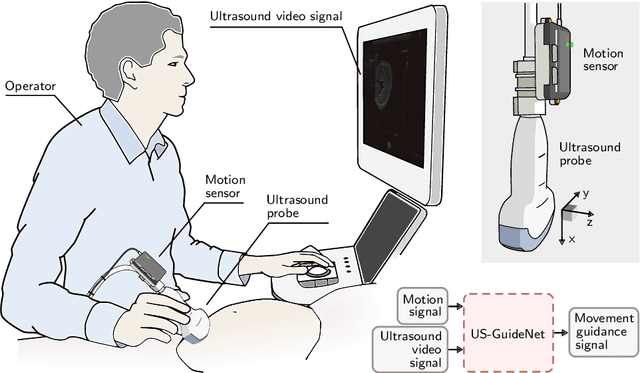
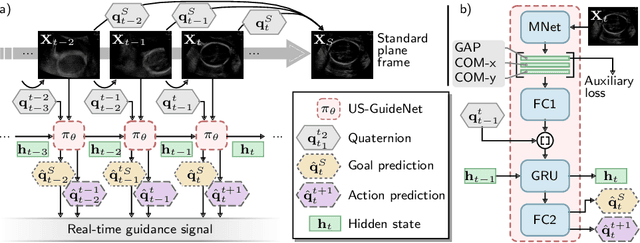
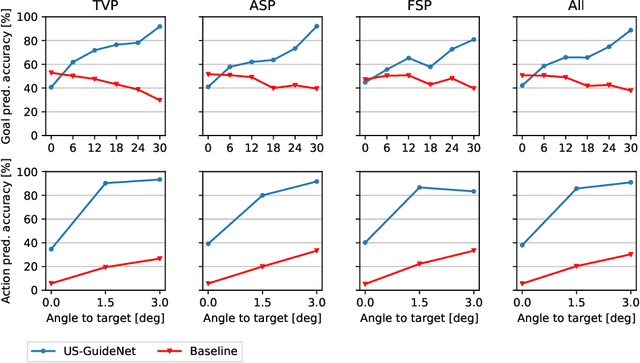
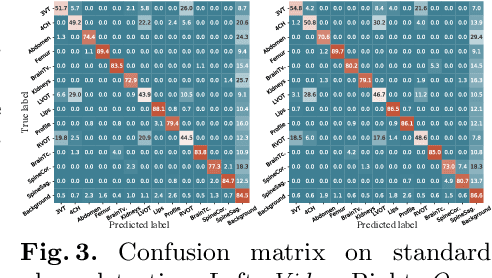
Abstract:We present the first system that provides real-time probe movement guidance for acquiring standard planes in routine freehand obstetric ultrasound scanning. Such a system can contribute to the worldwide deployment of obstetric ultrasound scanning by lowering the required level of operator expertise. The system employs an artificial neural network that receives the ultrasound video signal and the motion signal of an inertial measurement unit (IMU) that is attached to the probe, and predicts a guidance signal. The network termed US-GuideNet predicts either the movement towards the standard plane position (goal prediction), or the next movement that an expert sonographer would perform (action prediction). While existing models for other ultrasound applications are trained with simulations or phantoms, we train our model with real-world ultrasound video and probe motion data from 464 routine clinical scans by 17 accredited sonographers. Evaluations for 3 standard plane types show that the model provides a useful guidance signal with an accuracy of 88.8% for goal prediction and 90.9% for action prediction.
Unified Image and Video Saliency Modeling
Mar 11, 2020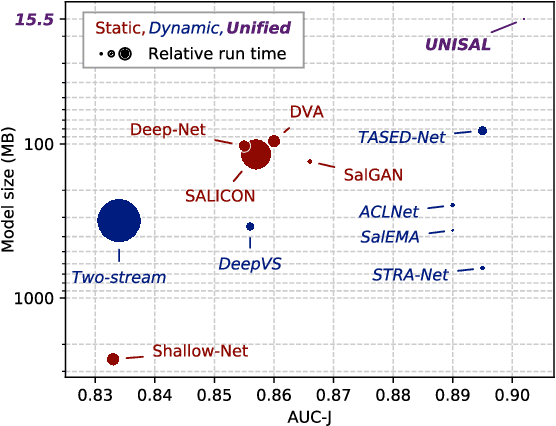
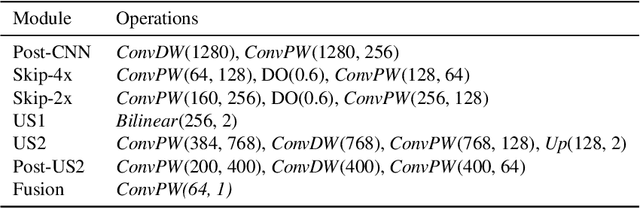
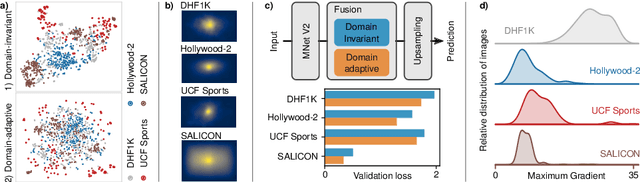
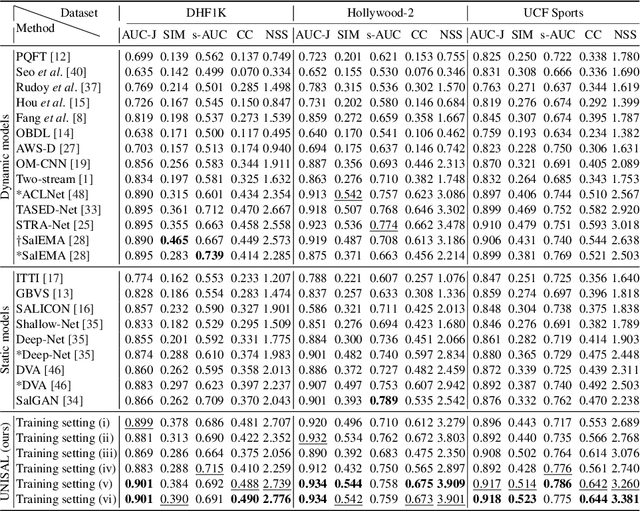
Abstract:Visual saliency modeling for images and videos is treated as two independent tasks in recent computer vision literature. On the one hand, image saliency modeling is a well-studied problem and progress on benchmarks like \mbox{SALICON} and MIT300 is slowing. For video saliency prediction on the other hand, rapid gains have been achieved on the recent DHF1K benchmark through network architectures that are optimized for this task. Here, we take a step back and ask: Can image and video saliency modeling be approached via a unified model, with mutual benefit? We find that it is crucial to model the domain shift between image and video saliency data and between different video saliency datasets for effective joint modeling. We identify different sources of domain shift and address them through four novel domain adaptation techniques - Domain-Adaptive Priors, Domain-Adaptive Fusion, Domain-Adaptive Smoothing and Bypass-RNN - in addition to an improved formulation of learned Gaussian priors. We integrate these techniques into a simple and lightweight encoder-RNN-decoder-style network, UNISAL, and train the entire network simultaneously with image and video saliency data. We evaluate our method on the video saliency datasets DHF1K, Hollywood-2 and UCF-Sports, as well as the image saliency datasets SALICON and MIT300. With one set of parameters, our method achieves state-of-the-art performance on all video saliency datasets and is on par with the state-of-the-art for image saliency prediction, despite a 5 to 20-fold reduction in model size and the fastest runtime among all competing deep models. We provide retrospective analyses and ablation studies which demonstrate the importance of the domain shift modeling. The code is available at https://github.com/rdroste/unisal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge