J. Alison Noble
Self-supervised Normality Learning and Divergence Vector-guided Model Merging for Zero-shot Congenital Heart Disease Detection in Fetal Ultrasound Videos
Mar 10, 2025Abstract:Congenital Heart Disease (CHD) is one of the leading causes of fetal mortality, yet the scarcity of labeled CHD data and strict privacy regulations surrounding fetal ultrasound (US) imaging present significant challenges for the development of deep learning-based models for CHD detection. Centralised collection of large real-world datasets for rare conditions, such as CHD, from large populations requires significant co-ordination and resource. In addition, data governance rules increasingly prevent data sharing between sites. To address these challenges, we introduce, for the first time, a novel privacy-preserving, zero-shot CHD detection framework that formulates CHD detection as a normality modeling problem integrated with model merging. In our framework dubbed Sparse Tube Ultrasound Distillation (STUD), each hospital site first trains a sparse video tube-based self-supervised video anomaly detection (VAD) model on normal fetal heart US clips with self-distillation loss. This enables site-specific models to independently learn the distribution of healthy cases. To aggregate knowledge across the decentralized models while maintaining privacy, we propose a Divergence Vector-Guided Model Merging approach, DivMerge, that combines site-specific models into a single VAD model without data exchange. Our approach preserves domain-agnostic rich spatio-temporal representations, ensuring generalization to unseen CHD cases. We evaluated our approach on real-world fetal US data collected from 5 hospital sites. Our merged model outperformed site-specific models by 23.77% and 30.13% in accuracy and F1-score respectively on external test sets.
FedPIA -- Permuting and Integrating Adapters leveraging Wasserstein Barycenters for Finetuning Foundation Models in Multi-Modal Federated Learning
Dec 19, 2024Abstract:Large Vision-Language Models typically require large text and image datasets for effective fine-tuning. However, collecting data from various sites, especially in healthcare, is challenging due to strict privacy regulations. An alternative is to fine-tune these models on end-user devices, such as in medical clinics, without sending data to a server. These local clients typically have limited computing power and small datasets, which are not enough for fully fine-tuning large VLMs on their own. A naive solution to these scenarios is to leverage parameter-efficient fine-tuning (PEFT) strategies and apply federated learning (FL) algorithms to combine the learned adapter weights, thereby respecting the resource limitations and data privacy. However, this approach does not fully leverage the knowledge from multiple adapters trained on diverse data distributions and for diverse tasks. The adapters are adversely impacted by data heterogeneity and task heterogeneity across clients resulting in suboptimal convergence. To this end, we propose a novel framework called FedPIA that improves upon the naive combinations of FL and PEFT by introducing Permutation and Integration of the local Adapters in the server and global Adapters in the clients exploiting Wasserstein barycenters for improved blending of client-specific and client-agnostic knowledge. This layerwise permutation helps to bridge the gap in the parameter space of local and global adapters before integration. We conduct over 2000 client-level experiments utilizing 48 medical image datasets across five different medical vision-language FL task settings encompassing visual question answering as well as image and report-based multi-label disease detection. Our experiments involving diverse client settings, ten different modalities, and two VLM backbones demonstrate that FedPIA consistently outperforms the state-of-the-art PEFT-FL baselines.
F$^3$OCUS -- Federated Finetuning of Vision-Language Foundation Models with Optimal Client Layer Updating Strategy via Multi-objective Meta-Heuristics
Nov 17, 2024



Abstract:Effective training of large Vision-Language Models (VLMs) on resource-constrained client devices in Federated Learning (FL) requires the usage of parameter-efficient fine-tuning (PEFT) strategies. To this end, we demonstrate the impact of two factors \textit{viz.}, client-specific layer importance score that selects the most important VLM layers for fine-tuning and inter-client layer diversity score that encourages diverse layer selection across clients for optimal VLM layer selection. We first theoretically motivate and leverage the principal eigenvalue magnitude of layerwise Neural Tangent Kernels and show its effectiveness as client-specific layer importance score. Next, we propose a novel layer updating strategy dubbed F$^3$OCUS that jointly optimizes the layer importance and diversity factors by employing a data-free, multi-objective, meta-heuristic optimization on the server. We explore 5 different meta-heuristic algorithms and compare their effectiveness for selecting model layers and adapter layers towards PEFT-FL. Furthermore, we release a new MedVQA-FL dataset involving overall 707,962 VQA triplets and 9 modality-specific clients and utilize it to train and evaluate our method. Overall, we conduct more than 10,000 client-level experiments on 6 Vision-Language FL task settings involving 58 medical image datasets and 4 different VLM architectures of varying sizes to demonstrate the effectiveness of the proposed method.
Pose-GuideNet: Automatic Scanning Guidance for Fetal Head Ultrasound from Pose Estimation
Aug 19, 2024



Abstract:3D pose estimation from a 2D cross-sectional view enables healthcare professionals to navigate through the 3D space, and such techniques initiate automatic guidance in many image-guided radiology applications. In this work, we investigate how estimating 3D fetal pose from freehand 2D ultrasound scanning can guide a sonographer to locate a head standard plane. Fetal head pose is estimated by the proposed Pose-GuideNet, a novel 2D/3D registration approach to align freehand 2D ultrasound to a 3D anatomical atlas without the acquisition of 3D ultrasound. To facilitate the 2D to 3D cross-dimensional projection, we exploit the prior knowledge in the atlas to align the standard plane frame in a freehand scan. A semantic-aware contrastive-based approach is further proposed to align the frames that are off standard planes based on their anatomical similarity. In the experiment, we enhance the existing assessment of freehand image localization by comparing the transformation of its estimated pose towards standard plane with the corresponding probe motion, which reflects the actual view change in 3D anatomy. Extensive results on two clinical head biometry tasks show that Pose-GuideNet not only accurately predicts pose but also successfully predicts the direction of the fetal head. Evaluations with probe motions further demonstrate the feasibility of adopting Pose-GuideNet for freehand ultrasound-assisted navigation in a sensor-free environment.
TextCAVs: Debugging vision models using text
Aug 16, 2024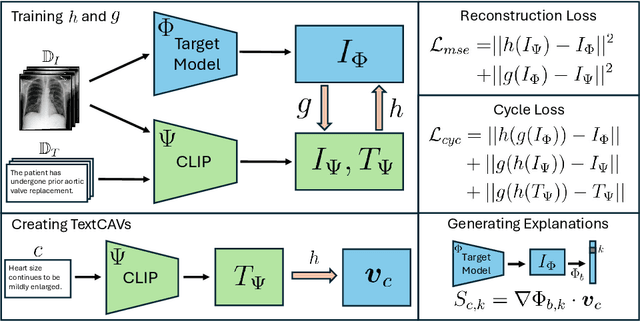
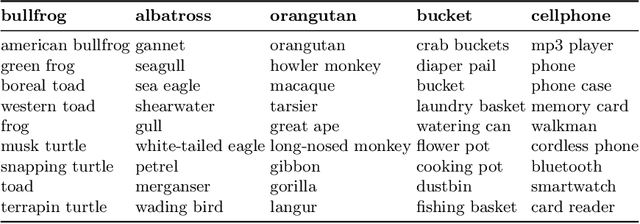
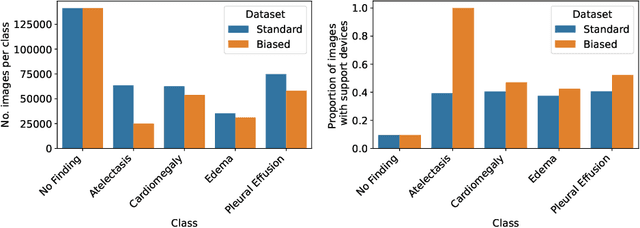
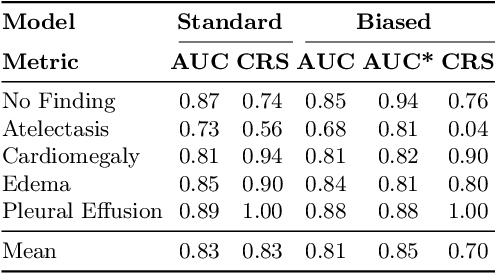
Abstract:Concept-based interpretability methods are a popular form of explanation for deep learning models which provide explanations in the form of high-level human interpretable concepts. These methods typically find concept activation vectors (CAVs) using a probe dataset of concept examples. This requires labelled data for these concepts -- an expensive task in the medical domain. We introduce TextCAVs: a novel method which creates CAVs using vision-language models such as CLIP, allowing for explanations to be created solely using text descriptions of the concept, as opposed to image exemplars. This reduced cost in testing concepts allows for many concepts to be tested and for users to interact with the model, testing new ideas as they are thought of, rather than a delay caused by image collection and annotation. In early experimental results, we demonstrate that TextCAVs produces reasonable explanations for a chest x-ray dataset (MIMIC-CXR) and natural images (ImageNet), and that these explanations can be used to debug deep learning-based models.
MMSummary: Multimodal Summary Generation for Fetal Ultrasound Video
Aug 07, 2024



Abstract:We present the first automated multimodal summary generation system, MMSummary, for medical imaging video, particularly with a focus on fetal ultrasound analysis. Imitating the examination process performed by a human sonographer, MMSummary is designed as a three-stage pipeline, progressing from keyframe detection to keyframe captioning and finally anatomy segmentation and measurement. In the keyframe detection stage, an innovative automated workflow is proposed to progressively select a concise set of keyframes, preserving sufficient video information without redundancy. Subsequently, we adapt a large language model to generate meaningful captions for fetal ultrasound keyframes in the keyframe captioning stage. If a keyframe is captioned as fetal biometry, the segmentation and measurement stage estimates biometric parameters by segmenting the region of interest according to the textual prior. The MMSummary system provides comprehensive summaries for fetal ultrasound examinations and based on reported experiments is estimated to reduce scanning time by approximately 31.5%, thereby suggesting the potential to enhance clinical workflow efficiency.
Feasibility of Federated Learning from Client Databases with Different Brain Diseases and MRI Modalities
Jun 17, 2024



Abstract:Segmentation models for brain lesions in MRI are commonly developed for a specific disease and trained on data with a predefined set of MRI modalities. Each such model cannot segment the disease using data with a different set of MRI modalities, nor can it segment any other type of disease. Moreover, this training paradigm does not allow a model to benefit from learning from heterogeneous databases that may contain scans and segmentation labels for different types of brain pathologies and diverse sets of MRI modalities. Is it feasible to use Federated Learning (FL) for training a single model on client databases that contain scans and labels of different brain pathologies and diverse sets of MRI modalities? We demonstrate promising results by combining appropriate, simple, and practical modifications to the model and training strategy: Designing a model with input channels that cover the whole set of modalities available across clients, training with random modality drop, and exploring the effects of feature normalization methods. Evaluation on 7 brain MRI databases with 5 different diseases shows that such FL framework can train a single model that is shown to be very promising in segmenting all disease types seen during training. Importantly, it is able to segment these diseases in new databases that contain sets of modalities different from those in training clients. These results demonstrate, for the first time, feasibility and effectiveness of using FL to train a single segmentation model on decentralised data with diverse brain diseases and MRI modalities, a necessary step towards leveraging heterogeneous real-world databases. Code will be made available at: https://github.com/FelixWag/FL-MultiDisease-MRI
IterMask2: Iterative Unsupervised Anomaly Segmentation via Spatial and Frequency Masking for Brain Lesions in MRI
Jun 04, 2024Abstract:Unsupervised anomaly segmentation approaches to pathology segmentation train a model on images of healthy subjects, that they define as the 'normal' data distribution. At inference, they aim to segment any pathologies in new images as 'anomalies', as they exhibit patterns that deviate from those in 'normal' training data. Prevailing methods follow the 'corrupt-and-reconstruct' paradigm. They intentionally corrupt an input image, reconstruct it to follow the learned 'normal' distribution, and subsequently segment anomalies based on reconstruction error. Corrupting an input image, however, inevitably leads to suboptimal reconstruction even of normal regions, causing false positives. To alleviate this, we propose a novel iterative spatial mask-refining strategy IterMask2. We iteratively mask areas of the image, reconstruct them, and update the mask based on reconstruction error. This iterative process progressively adds information about areas that are confidently normal as per the model. The increasing content guides reconstruction of nearby masked areas, improving reconstruction of normal tissue under these areas, reducing false positives. We also use high-frequency image content as an auxiliary input to provide additional structural information for masked areas. This further improves reconstruction error of normal in comparison to anomalous areas, facilitating segmentation of the latter. We conduct experiments on several brain lesion datasets and demonstrate effectiveness of our method. Code is available at: https://github.com/ZiyunLiang/IterMasks2
Explaining Explainability: Understanding Concept Activation Vectors
Apr 04, 2024Abstract:Recent interpretability methods propose using concept-based explanations to translate the internal representations of deep learning models into a language that humans are familiar with: concepts. This requires understanding which concepts are present in the representation space of a neural network. One popular method for finding concepts is Concept Activation Vectors (CAVs), which are learnt using a probe dataset of concept exemplars. In this work, we investigate three properties of CAVs. CAVs may be: (1) inconsistent between layers, (2) entangled with different concepts, and (3) spatially dependent. Each property provides both challenges and opportunities in interpreting models. We introduce tools designed to detect the presence of these properties, provide insight into how they affect the derived explanations, and provide recommendations to minimise their impact. Understanding these properties can be used to our advantage. For example, we introduce spatially dependent CAVs to test if a model is translation invariant with respect to a specific concept and class. Our experiments are performed on ImageNet and a new synthetic dataset, Elements. Elements is designed to capture a known ground truth relationship between concepts and classes. We release this dataset to facilitate further research in understanding and evaluating interpretability methods.
Semi-weakly-supervised neural network training for medical image registration
Feb 16, 2024



Abstract:For training registration networks, weak supervision from segmented corresponding regions-of-interest (ROIs) have been proven effective for (a) supplementing unsupervised methods, and (b) being used independently in registration tasks in which unsupervised losses are unavailable or ineffective. This correspondence-informing supervision entails cost in annotation that requires significant specialised effort. This paper describes a semi-weakly-supervised registration pipeline that improves the model performance, when only a small corresponding-ROI-labelled dataset is available, by exploiting unlabelled image pairs. We examine two types of augmentation methods by perturbation on network weights and image resampling, such that consistency-based unsupervised losses can be applied on unlabelled data. The novel WarpDDF and RegCut approaches are proposed to allow commutative perturbation between an image pair and the predicted spatial transformation (i.e. respective input and output of registration networks), distinct from existing perturbation methods for classification or segmentation. Experiments using 589 male pelvic MR images, labelled with eight anatomical ROIs, show the improvement in registration performance and the ablated contributions from the individual strategies. Furthermore, this study attempts to construct one of the first computational atlases for pelvic structures, enabled by registering inter-subject MRs, and quantifies the significant differences due to the proposed semi-weak supervision with a discussion on the potential clinical use of example atlas-derived statistics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge