Imanol Luengo
Learning from Single Timestamps: Complexity Estimation in Laparoscopic Cholecystectomy
Nov 06, 2025Abstract:Purpose: Accurate assessment of surgical complexity is essential in Laparoscopic Cholecystectomy (LC), where severe inflammation is associated with longer operative times and increased risk of postoperative complications. The Parkland Grading Scale (PGS) provides a clinically validated framework for stratifying inflammation severity; however, its automation in surgical videos remains largely unexplored, particularly in realistic scenarios where complete videos must be analyzed without prior manual curation. Methods: In this work, we introduce STC-Net, a novel framework for SingleTimestamp-based Complexity estimation in LC via the PGS, designed to operate under weak temporal supervision. Unlike prior methods limited to static images or manually trimmed clips, STC-Net operates directly on full videos. It jointly performs temporal localization and grading through a localization, window proposal, and grading module. We introduce a novel loss formulation combining hard and soft localization objectives and background-aware grading supervision. Results: Evaluated on a private dataset of 1,859 LC videos, STC-Net achieves an accuracy of 62.11% and an F1-score of 61.42%, outperforming non-localized baselines by over 10% in both metrics and highlighting the effectiveness of weak supervision for surgical complexity assessment. Conclusion: STC-Net demonstrates a scalable and effective approach for automated PGS-based surgical complexity estimation from full LC videos, making it promising for post-operative analysis and surgical training.
Comparative validation of surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation in endoscopy: Results of the PhaKIR 2024 challenge
Jul 22, 2025Abstract:Reliable recognition and localization of surgical instruments in endoscopic video recordings are foundational for a wide range of applications in computer- and robot-assisted minimally invasive surgery (RAMIS), including surgical training, skill assessment, and autonomous assistance. However, robust performance under real-world conditions remains a significant challenge. Incorporating surgical context - such as the current procedural phase - has emerged as a promising strategy to improve robustness and interpretability. To address these challenges, we organized the Surgical Procedure Phase, Keypoint, and Instrument Recognition (PhaKIR) sub-challenge as part of the Endoscopic Vision (EndoVis) challenge at MICCAI 2024. We introduced a novel, multi-center dataset comprising thirteen full-length laparoscopic cholecystectomy videos collected from three distinct medical institutions, with unified annotations for three interrelated tasks: surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation. Unlike existing datasets, ours enables joint investigation of instrument localization and procedural context within the same data while supporting the integration of temporal information across entire procedures. We report results and findings in accordance with the BIAS guidelines for biomedical image analysis challenges. The PhaKIR sub-challenge advances the field by providing a unique benchmark for developing temporally aware, context-driven methods in RAMIS and offers a high-quality resource to support future research in surgical scene understanding.
A spatio-temporal network for video semantic segmentation in surgical videos
Jun 19, 2023Abstract:Semantic segmentation in surgical videos has applications in intra-operative guidance, post-operative analytics and surgical education. Segmentation models need to provide accurate and consistent predictions since temporally inconsistent identification of anatomical structures can impair usability and hinder patient safety. Video information can alleviate these challenges leading to reliable models suitable for clinical use. We propose a novel architecture for modelling temporal relationships in videos. The proposed model includes a spatio-temporal decoder to enable video semantic segmentation by improving temporal consistency across frames. The encoder processes individual frames whilst the decoder processes a temporal batch of adjacent frames. The proposed decoder can be used on top of any segmentation encoder to improve temporal consistency. Model performance was evaluated on the CholecSeg8k dataset and a private dataset of robotic Partial Nephrectomy procedures. Segmentation performance was improved when the temporal decoder was applied across both datasets. The proposed model also displayed improvements in temporal consistency.
Self-Knowledge Distillation for Surgical Phase Recognition
Jun 15, 2023



Abstract:Purpose: Advances in surgical phase recognition are generally led by training deeper networks. Rather than going further with a more complex solution, we believe that current models can be exploited better. We propose a self-knowledge distillation framework that can be integrated into current state-of-the-art (SOTA) models without requiring any extra complexity to the models or annotations. Methods: Knowledge distillation is a framework for network regularization where knowledge is distilled from a teacher network to a student network. In self-knowledge distillation, the student model becomes the teacher such that the network learns from itself. Most phase recognition models follow an encoder-decoder framework. Our framework utilizes self-knowledge distillation in both stages. The teacher model guides the training process of the student model to extract enhanced feature representations from the encoder and build a more robust temporal decoder to tackle the over-segmentation problem. Results: We validate our proposed framework on the public dataset Cholec80. Our framework is embedded on top of four popular SOTA approaches and consistently improves their performance. Specifically, our best GRU model boosts performance by +3.33% accuracy and +3.95% F1-score over the same baseline model. Conclusion: We embed a self-knowledge distillation framework for the first time in the surgical phase recognition training pipeline. Experimental results demonstrate that our simple yet powerful framework can improve performance of existing phase recognition models. Moreover, our extensive experiments show that even with 75% of the training set we still achieve performance on par with the same baseline model trained on the full set.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
CholecTriplet2021: A benchmark challenge for surgical action triplet recognition
Apr 10, 2022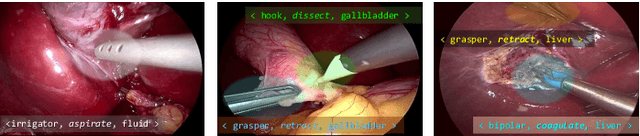
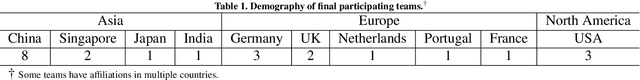
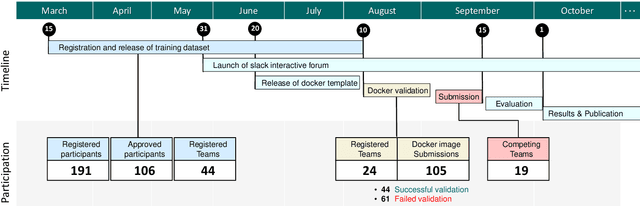
Abstract:Context-aware decision support in the operating room can foster surgical safety and efficiency by leveraging real-time feedback from surgical workflow analysis. Most existing works recognize surgical activities at a coarse-grained level, such as phases, steps or events, leaving out fine-grained interaction details about the surgical activity; yet those are needed for more helpful AI assistance in the operating room. Recognizing surgical actions as triplets of <instrument, verb, target> combination delivers comprehensive details about the activities taking place in surgical videos. This paper presents CholecTriplet2021: an endoscopic vision challenge organized at MICCAI 2021 for the recognition of surgical action triplets in laparoscopic videos. The challenge granted private access to the large-scale CholecT50 dataset, which is annotated with action triplet information. In this paper, we present the challenge setup and assessment of the state-of-the-art deep learning methods proposed by the participants during the challenge. A total of 4 baseline methods from the challenge organizers and 19 new deep learning algorithms by competing teams are presented to recognize surgical action triplets directly from surgical videos, achieving mean average precision (mAP) ranging from 4.2% to 38.1%. This study also analyzes the significance of the results obtained by the presented approaches, performs a thorough methodological comparison between them, in-depth result analysis, and proposes a novel ensemble method for enhanced recognition. Our analysis shows that surgical workflow analysis is not yet solved, and also highlights interesting directions for future research on fine-grained surgical activity recognition which is of utmost importance for the development of AI in surgery.
2020 CATARACTS Semantic Segmentation Challenge
Oct 21, 2021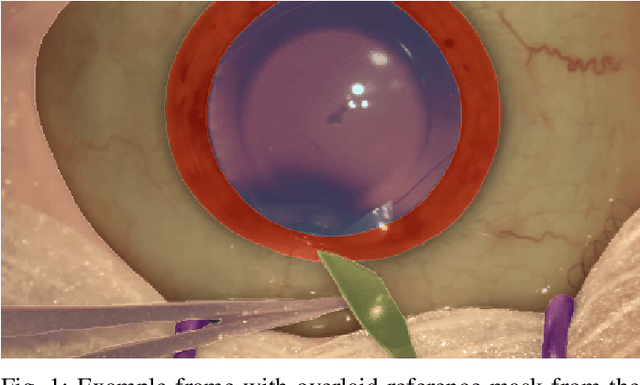
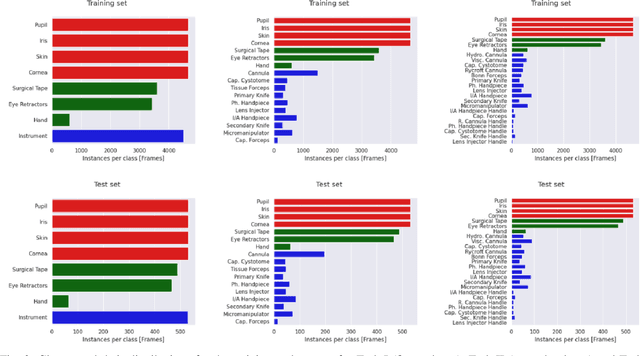
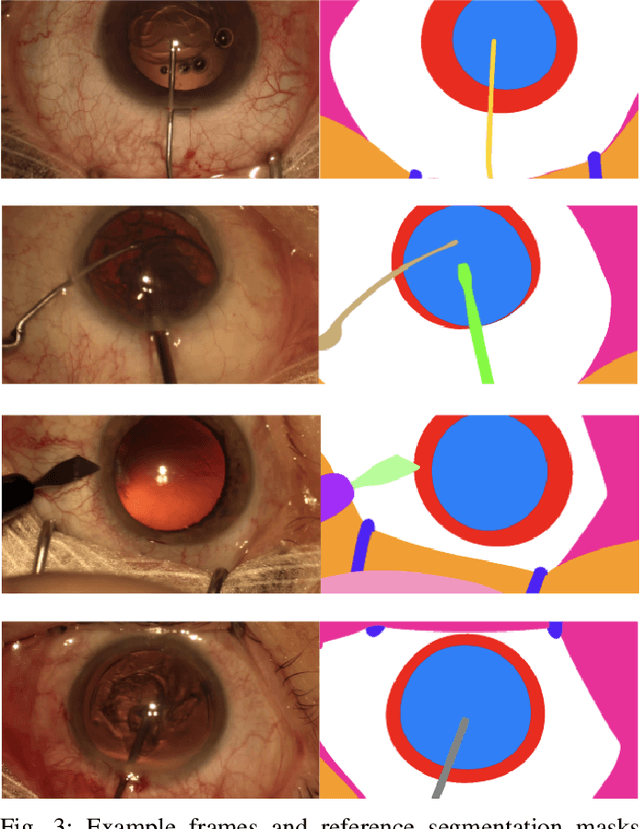
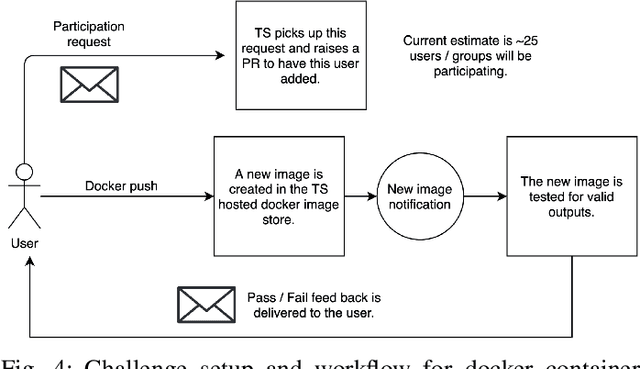
Abstract:Surgical scene segmentation is essential for anatomy and instrument localization which can be further used to assess tissue-instrument interactions during a surgical procedure. In 2017, the Challenge on Automatic Tool Annotation for cataRACT Surgery (CATARACTS) released 50 cataract surgery videos accompanied by instrument usage annotations. These annotations included frame-level instrument presence information. In 2020, we released pixel-wise semantic annotations for anatomy and instruments for 4670 images sampled from 25 videos of the CATARACTS training set. The 2020 CATARACTS Semantic Segmentation Challenge, which was a sub-challenge of the 2020 MICCAI Endoscopic Vision (EndoVis) Challenge, presented three sub-tasks to assess participating solutions on anatomical structure and instrument segmentation. Their performance was assessed on a hidden test set of 531 images from 10 videos of the CATARACTS test set.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020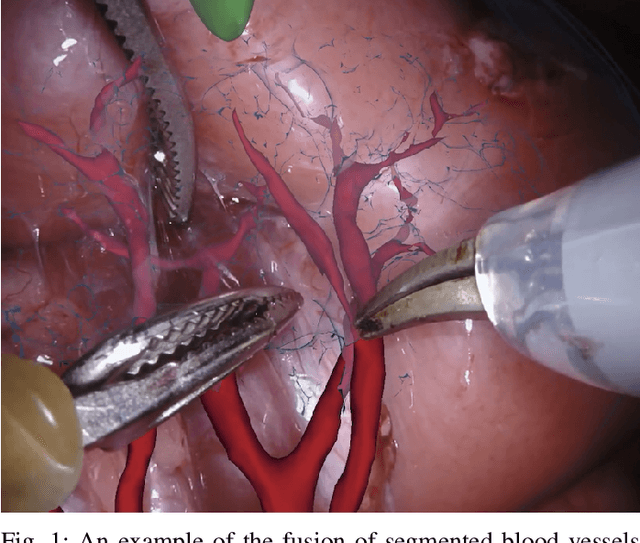
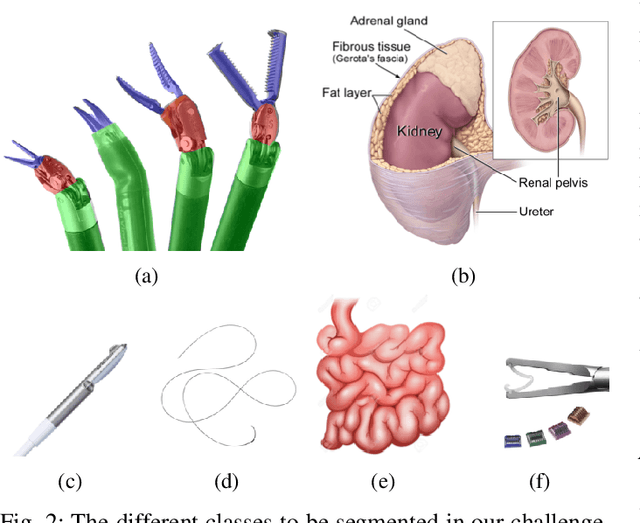
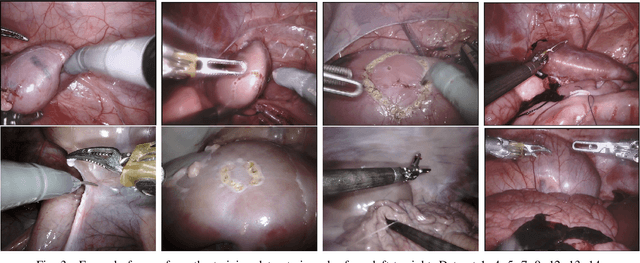
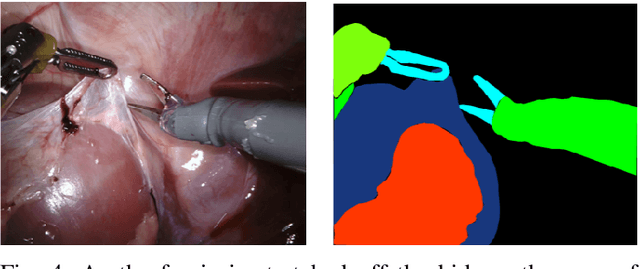
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
CaDIS: Cataract Dataset for Image Segmentation
Jul 19, 2019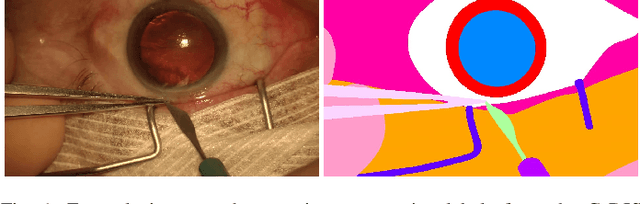
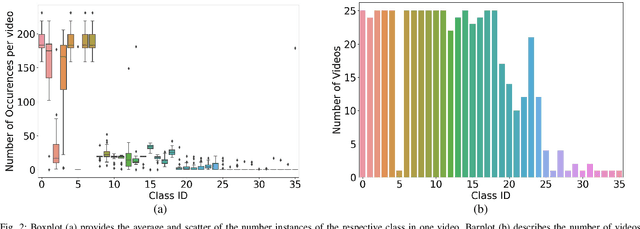
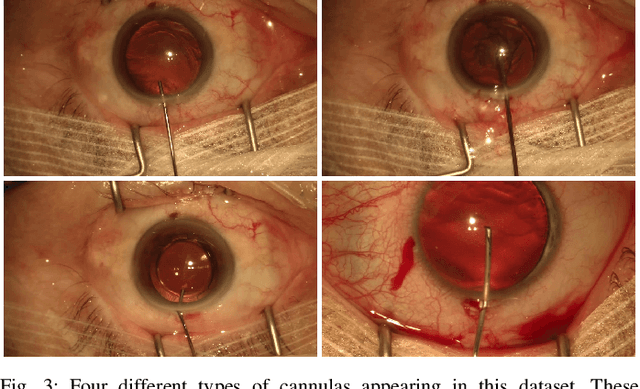
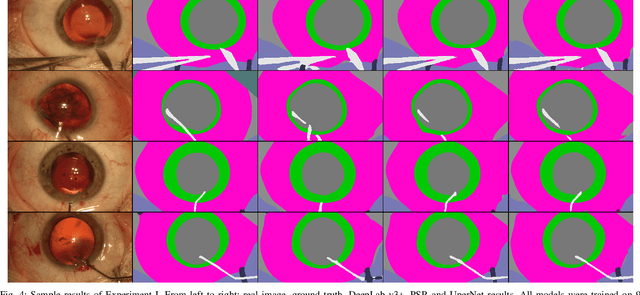
Abstract:Video signals provide a wealth of information about surgical procedures and are the main sensory cue for surgeons. Video processing and understanding can be used to empower computer assisted interventions (CAI) as well as the development of detailed post-operative analysis of the surgical intervention. A fundamental building block to such capabilities is the ability to understand and segment video into semantic labels that differentiate and localize tissue types and different instruments. Deep learning has advanced semantic segmentation techniques dramatically in recent years but is fundamentally reliant on the availability of labelled datasets used to train models. In this paper, we introduce a high quality dataset for semantic segmentation in Cataract surgery. We generated this dataset from the CATARACTS challenge dataset, which is publicly available. To the best of our knowledge, this dataset has the highest quality annotation in surgical data to date. We introduce the dataset and then show the automatic segmentation performance of state-of-the-art models on that dataset as a benchmark.
SurReal: enhancing Surgical simulation Realism using style transfer
Nov 07, 2018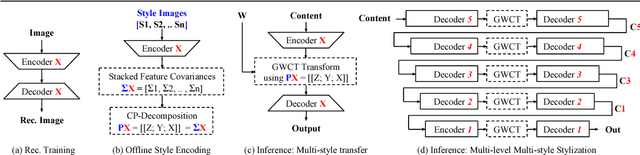
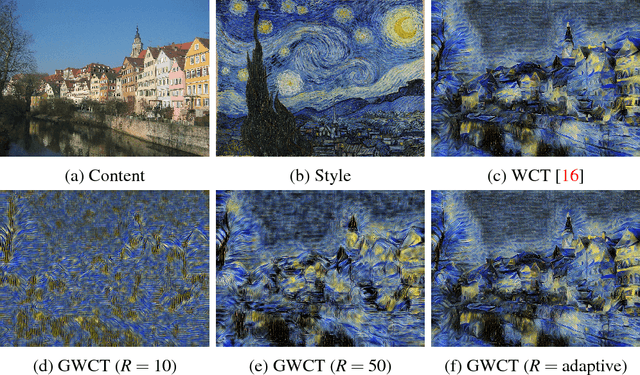
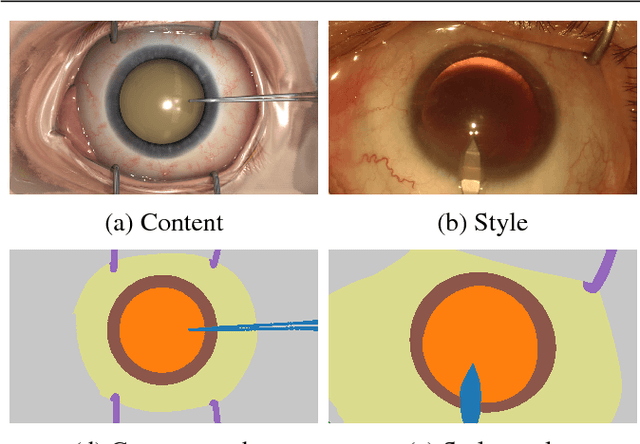
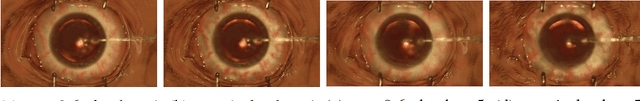
Abstract:Surgical simulation is an increasingly important element of surgical education. Using simulation can be a means to address some of the significant challenges in developing surgical skills with limited time and resources. The photo-realistic fidelity of simulations is a key feature that can improve the experience and transfer ratio of trainees. In this paper, we demonstrate how we can enhance the visual fidelity of existing surgical simulation by performing style transfer of multi-class labels from real surgical video onto synthetic content. We demonstrate our approach on simulations of cataract surgery using real data labels from an existing public dataset. Our results highlight the feasibility of the approach and also the powerful possibility to extend this technique to incorporate additional temporal constraints and to different applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge