Cheng Lei
Towards Real Zero-Shot Camouflaged Object Segmentation without Camouflaged Annotations
Oct 22, 2024Abstract:Camouflaged Object Segmentation (COS) faces significant challenges due to the scarcity of annotated data, where meticulous pixel-level annotation is both labor-intensive and costly, primarily due to the intricate object-background boundaries. Addressing the core question, "Can COS be effectively achieved in a zero-shot manner without manual annotations for any camouflaged object?" we affirmatively respond and introduce a robust zero-shot COS framework. This framework leverages the inherent local pattern bias of COS and employs a broad semantic feature space derived from salient object segmentation (SOS) for efficient zero-shot transfer. We incorporate an Masked Image Modeling (MIM) based image encoder optimized for Parameter-Efficient Fine-Tuning (PEFT), a Multimodal Large Language Model (M-LLM), and a Multi-scale Fine-grained Alignment (MFA) mechanism. The MIM pre-trained image encoder focuses on capturing essential low-level features, while the M-LLM generates caption embeddings processed alongside these visual cues. These embeddings are precisely aligned using MFA, enabling our framework to accurately interpret and navigate complex semantic contexts. To optimize operational efficiency, we introduce a learnable codebook that represents the M-LLM during inference, significantly reducing computational overhead. Our framework demonstrates its versatility and efficacy through rigorous experimentation, achieving state-of-the-art performance in zero-shot COS with $F_{\beta}^w$ scores of 72.9\% on CAMO and 71.7\% on COD10K. By removing the M-LLM during inference, we achieve an inference speed comparable to that of traditional end-to-end models, reaching 18.1 FPS. Code: https://github.com/R-LEI360725/ZSCOS-CaMF
iKUN: Speak to Trackers without Retraining
Dec 25, 2023Abstract:Referring multi-object tracking (RMOT) aims to track multiple objects based on input textual descriptions. Previous works realize it by simply integrating an extra textual module into the multi-object tracker. However, they typically need to retrain the entire framework and have difficulties in optimization. In this work, we propose an insertable Knowledge Unification Network, termed iKUN, to enable communication with off-the-shelf trackers in a plug-and-play manner. Concretely, a knowledge unification module (KUM) is designed to adaptively extract visual features based on textual guidance. Meanwhile, to improve the localization accuracy, we present a neural version of Kalman filter (NKF) to dynamically adjust process noise and observation noise based on the current motion status. Moreover, to address the problem of open-set long-tail distribution of textual descriptions, a test-time similarity calibration method is proposed to refine the confidence score with pseudo frequency. Extensive experiments on Refer-KITTI dataset verify the effectiveness of our framework. Finally, to speed up the development of RMOT, we also contribute a more challenging dataset, Refer-Dance, by extending public DanceTrack dataset with motion and dressing descriptions. The code and dataset will be released in https://github.com/dyhBUPT/iKUN.
Video-based Visible-Infrared Person Re-Identification with Auxiliary Samples
Nov 27, 2023Abstract:Visible-infrared person re-identification (VI-ReID) aims to match persons captured by visible and infrared cameras, allowing person retrieval and tracking in 24-hour surveillance systems. Previous methods focus on learning from cross-modality person images in different cameras. However, temporal information and single-camera samples tend to be neglected. To crack this nut, in this paper, we first contribute a large-scale VI-ReID dataset named BUPTCampus. Different from most existing VI-ReID datasets, it 1) collects tracklets instead of images to introduce rich temporal information, 2) contains pixel-aligned cross-modality sample pairs for better modality-invariant learning, 3) provides one auxiliary set to help enhance the optimization, in which each identity only appears in a single camera. Based on our constructed dataset, we present a two-stream framework as baseline and apply Generative Adversarial Network (GAN) to narrow the gap between the two modalities. To exploit the advantages introduced by the auxiliary set, we propose a curriculum learning based strategy to jointly learn from both primary and auxiliary sets. Moreover, we design a novel temporal k-reciprocal re-ranking method to refine the ranking list with fine-grained temporal correlation cues. Experimental results demonstrate the effectiveness of the proposed methods. We also reproduce 9 state-of-the-art image-based and video-based VI-ReID methods on BUPTCampus and our methods show substantial superiority to them. The codes and dataset are available at: https://github.com/dyhBUPT/BUPTCampus.
Adversarial Multiscale Feature Learning for Overlapping Chromosome Segmentation
Dec 22, 2020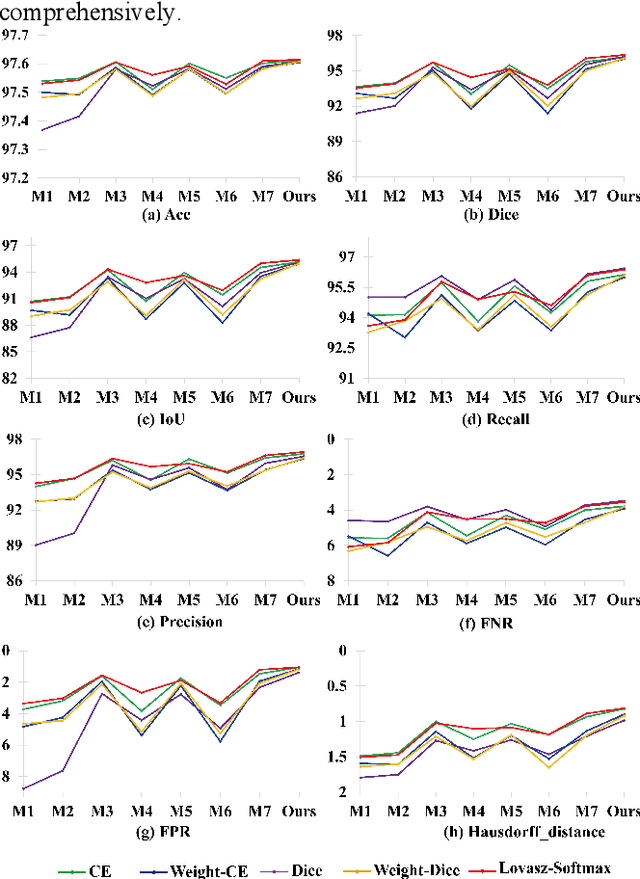
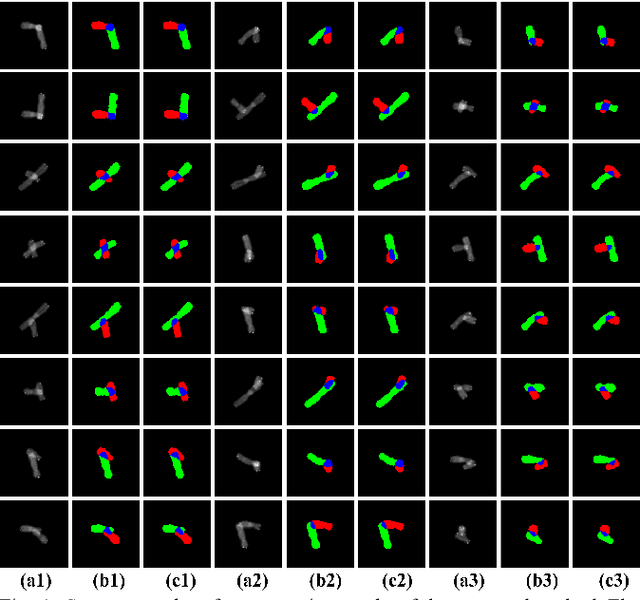

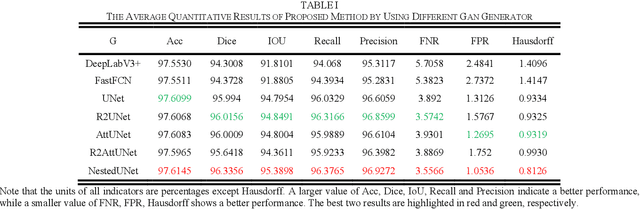
Abstract:Chromosome karyotype analysis is of great clinical importance in the diagnosis and treatment of diseases, especially for genetic diseases. Since manual analysis is highly time and effort consuming, computer-assisted automatic chromosome karyotype analysis based on images is routinely used to improve the efficiency and accuracy of the analysis. Due to the strip shape of the chromosomes, they easily get overlapped with each other when imaged, significantly affecting the accuracy of the analysis afterward. Conventional overlapping chromosome segmentation methods are usually based on manually tagged features, hence, the performance of which is easily affected by the quality, such as resolution and brightness, of the images. To address the problem, in this paper, we present an adversarial multiscale feature learning framework to improve the accuracy and adaptability of overlapping chromosome segmentation. Specifically, we first adopt the nested U-shape network with dense skip connections as the generator to explore the optimal representation of the chromosome images by exploiting multiscale features. Then we use the conditional generative adversarial network (cGAN) to generate images similar to the original ones, the training stability of which is enhanced by applying the least-square GAN objective. Finally, we employ Lovasz-Softmax to help the model converge in a continuous optimization setting. Comparing with the established algorithms, the performance of our framework is proven superior by using public datasets in eight evaluation criteria, showing its great potential in overlapping chromosome segmentation
A Preliminary Study on Data Augmentation of Deep Learning for Image Classification
Jun 09, 2019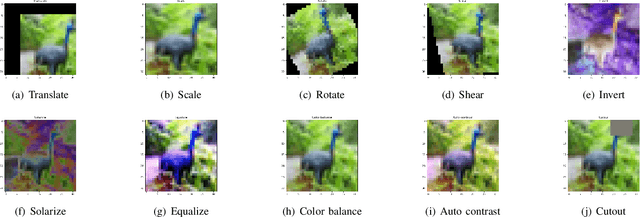
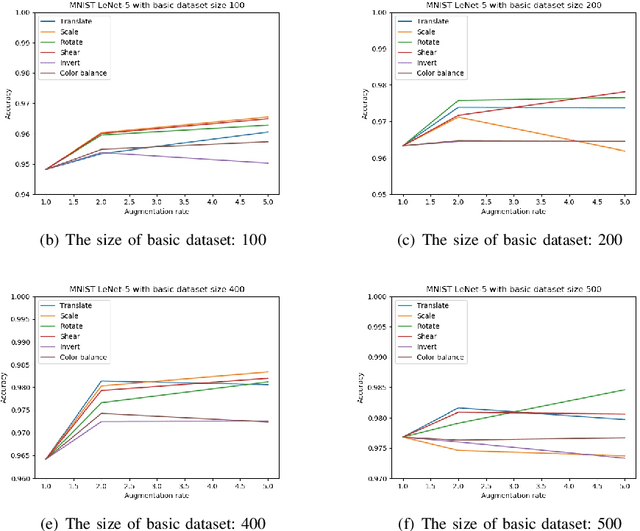
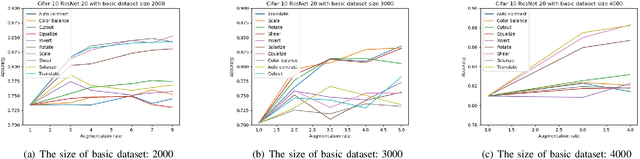
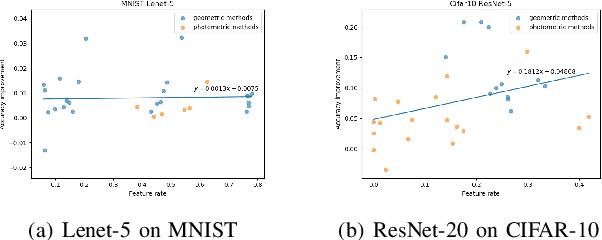
Abstract:Deep learning models have a large number of freeparameters that need to be calculated by effective trainingof the models on a great deal of training data to improvetheir generalization performance. However, data obtaining andlabeling is expensive in practice. Data augmentation is one of themethods to alleviate this problem. In this paper, we conduct apreliminary study on how three variables (augmentation method,augmentation rate and size of basic dataset per label) can affectthe accuracy of deep learning for image classification. The studyprovides some guidelines: (1) it is better to use transformationsthat alter the geometry of the images rather than those justlighting and color. (2) 2-3 times augmentation rate is good enoughfor training. (3) the smaller amount of data, the more obviouscontributions could have.
Exploring the Deep Feature Space of a Cell Classification Neural Network
Nov 15, 2018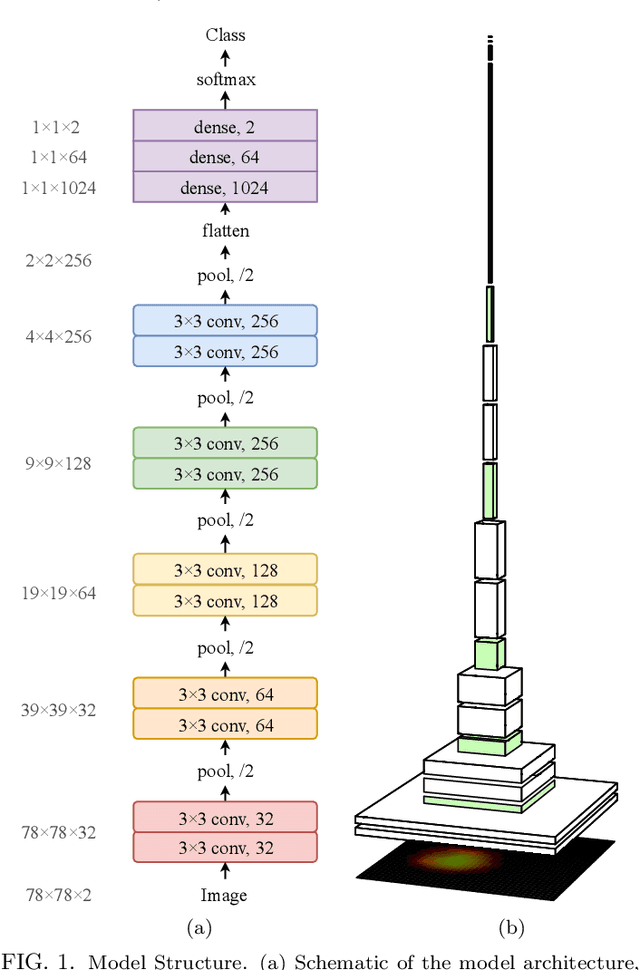
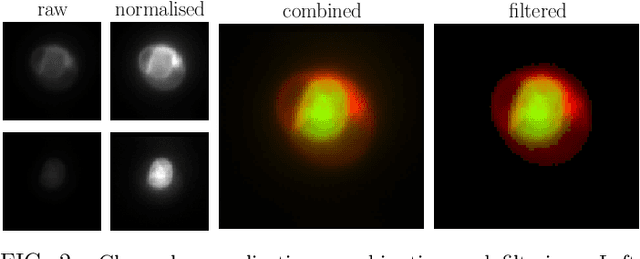
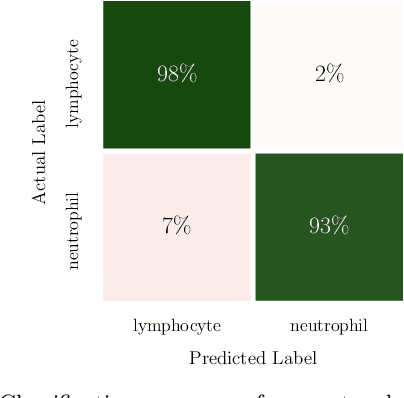
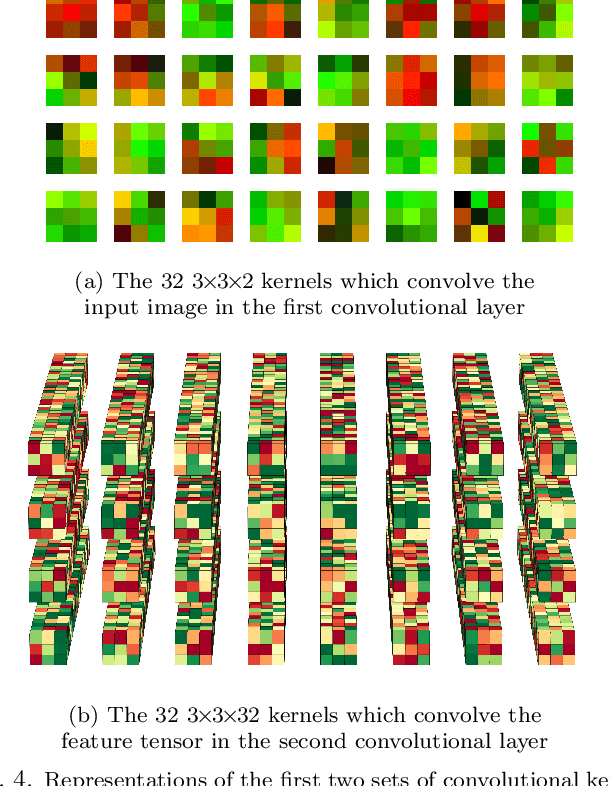
Abstract:In this paper, we present contemporary techniques for visualising the feature space of a deep learning image classification neural network. These techniques are viewed in the context of a feed-forward network trained to classify low resolution fluorescence images of white blood cells captured using optofluidic imaging. The model has two output classes corresponding to two different cell types, which are often difficult to distinguish by eye. This paper has two major sections. The first looks to develop the information space presented by dimension reduction techniques, such as t-SNE, used to embed high-dimensional pre-softmax layer activations into a two-dimensional plane. The second section looks at feature visualisation by optimisation to generate feature images representing the learned features of the network. Using and developing these techniques we visualise class separation and structures within the dataset at various depths using clustering algorithms and feature images; track the development of feature complexity as we ascend the network; and begin to extract the features the network has learnt by modulating single-channel feature images with up-scaled neuron activation maps to distinguish their most salient parts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge