Babak Ehteshami Bejnordi
Skip-Convolutions for Efficient Video Processing
Apr 23, 2021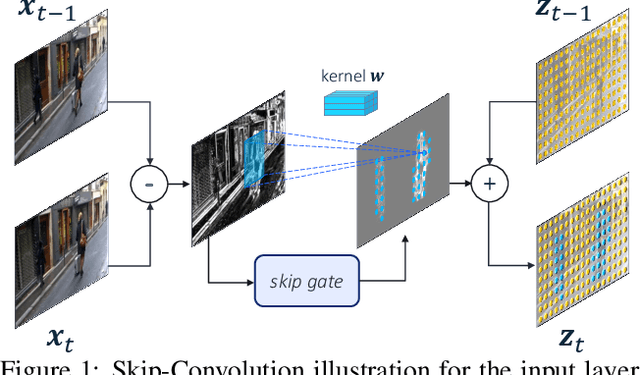
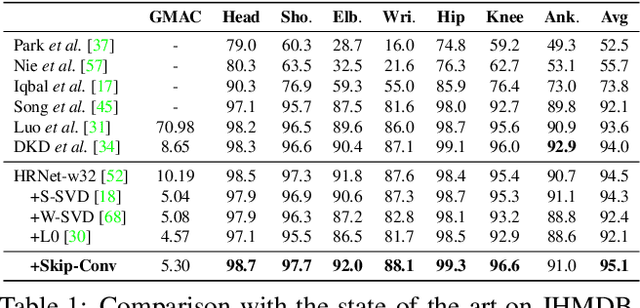
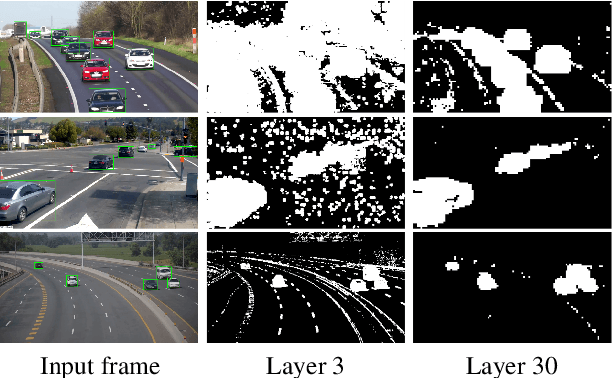
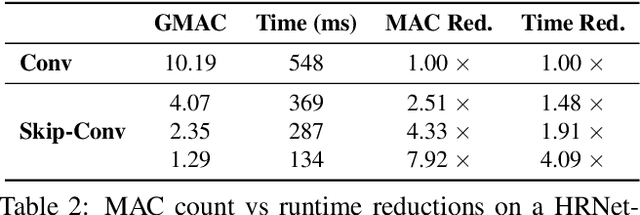
Abstract:We propose Skip-Convolutions to leverage the large amount of redundancies in video streams and save computations. Each video is represented as a series of changes across frames and network activations, denoted as residuals. We reformulate standard convolution to be efficiently computed on residual frames: each layer is coupled with a binary gate deciding whether a residual is important to the model prediction,~\eg foreground regions, or it can be safely skipped, e.g. background regions. These gates can either be implemented as an efficient network trained jointly with convolution kernels, or can simply skip the residuals based on their magnitude. Gating functions can also incorporate block-wise sparsity structures, as required for efficient implementation on hardware platforms. By replacing all convolutions with Skip-Convolutions in two state-of-the-art architectures, namely EfficientDet and HRNet, we reduce their computational cost consistently by a factor of 3~4x for two different tasks, without any accuracy drop. Extensive comparisons with existing model compression, as well as image and video efficiency methods demonstrate that Skip-Convolutions set a new state-of-the-art by effectively exploiting the temporal redundancies in videos.
TimeGate: Conditional Gating of Segments in Long-range Activities
Apr 03, 2020
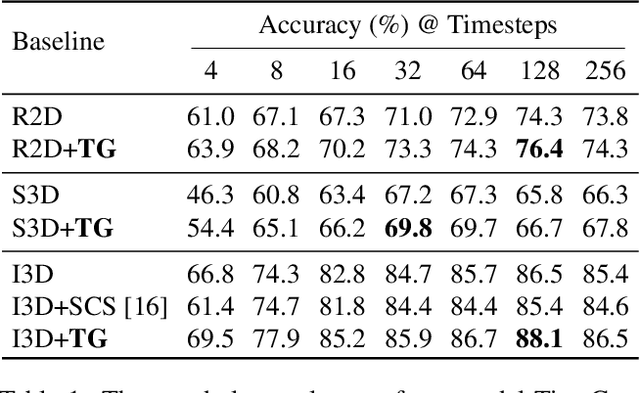
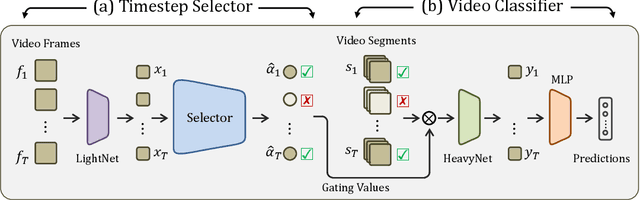
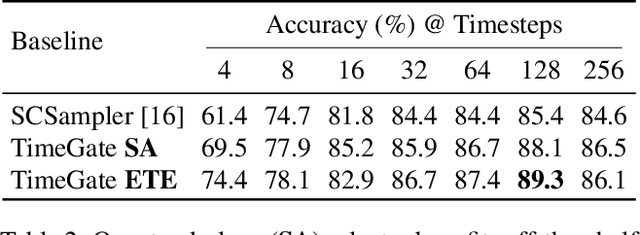
Abstract:When recognizing a long-range activity, exploring the entire video is exhaustive and computationally expensive, as it can span up to a few minutes. Thus, it is of great importance to sample only the salient parts of the video. We propose TimeGate, along with a novel conditional gating module, for sampling the most representative segments from the long-range activity. TimeGate has two novelties that address the shortcomings of previous sampling methods, as SCSampler. First, it enables a differentiable sampling of segments. Thus, TimeGate can be fitted with modern CNNs and trained end-to-end as a single and unified model.Second, the sampling is conditioned on both the segments and their context. Consequently, TimeGate is better suited for long-range activities, where the importance of a segment heavily depends on the video context.TimeGate reduces the computation of existing CNNs on three benchmarks for long-range activities: Charades, Breakfast and MultiThumos. In particular, TimeGate reduces the computation of I3D by 50% while maintaining the classification accuracy.
Conditional Channel Gated Networks for Task-Aware Continual Learning
Mar 31, 2020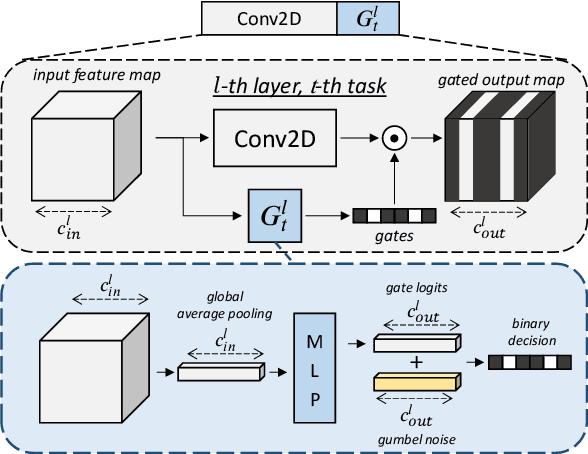
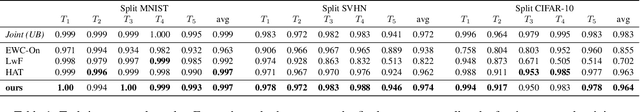
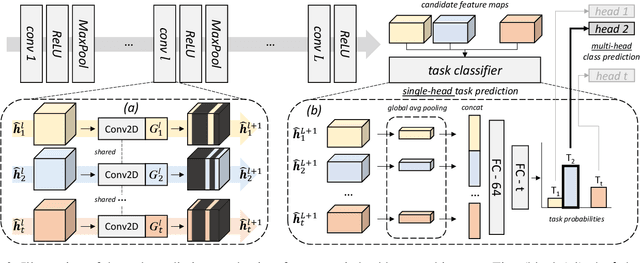
Abstract:Convolutional Neural Networks experience catastrophic forgetting when optimized on a sequence of learning problems: as they meet the objective of the current training examples, their performance on previous tasks drops drastically. In this work, we introduce a novel framework to tackle this problem with conditional computation. We equip each convolutional layer with task-specific gating modules, selecting which filters to apply on the given input. This way, we achieve two appealing properties. Firstly, the execution patterns of the gates allow to identify and protect important filters, ensuring no loss in the performance of the model for previously learned tasks. Secondly, by using a sparsity objective, we can promote the selection of a limited set of kernels, allowing to retain sufficient model capacity to digest new tasks.Existing solutions require, at test time, awareness of the task to which each example belongs to. This knowledge, however, may not be available in many practical scenarios. Therefore, we additionally introduce a task classifier that predicts the task label of each example, to deal with settings in which a task oracle is not available. We validate our proposal on four continual learning datasets. Results show that our model consistently outperforms existing methods both in the presence and the absence of a task oracle. Notably, on Split SVHN and Imagenet-50 datasets, our model yields up to 23.98% and 17.42% improvement in accuracy w.r.t. competing methods.
Batch-Shaped Channel Gated Networks
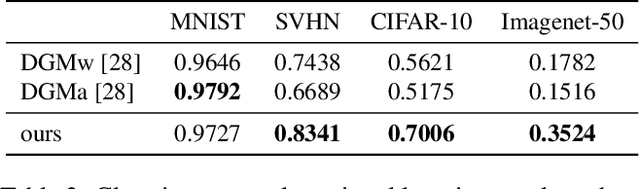
Jul 15, 2019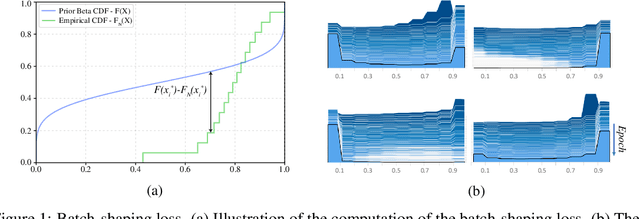
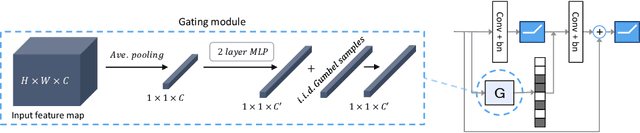
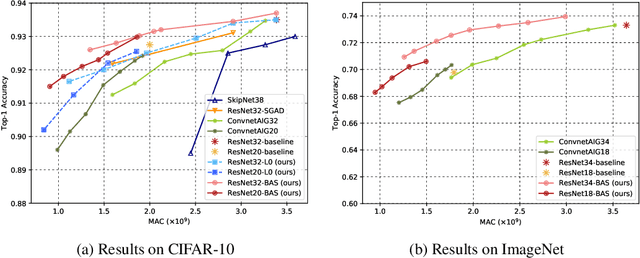
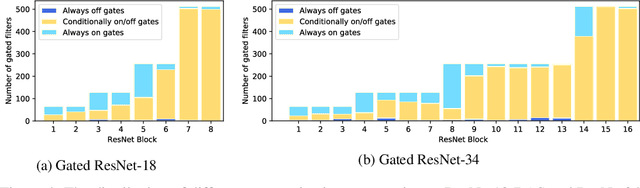
Abstract:We present a method for gating deep-learning architectures on a fine-grained level. Individual convolutional maps are turned on/off conditionally on features in the network. This method allows us to train neural networks with a large capacity, but lower inference time than the full network. To achieve this, we introduce a new residual block architecture that gates convolutional channels in a fine-grained manner. We also introduce a generally applicable tool "batch-shaping" that matches the marginal aggregate posteriors of features in a neural network to a pre-specified prior distribution. We use this novel technique to force gates to be more conditional on the data. We present results on CIFAR-10 and ImageNet datasets for image classification and Cityscapes for semantic segmentation. Our results show that our method can slim down large architectures conditionally, such that the average computational cost on the data is on par with a smaller architecture, but with higher accuracy. In particular, our ResNet34 gated network achieves a performance of 72.55% top-1 accuracy compared to the 69.76% accuracy of the baseline ResNet18 model, for similar complexity. We also show that the resulting networks automatically learn to use more features for difficult examples and fewer features for simple examples.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018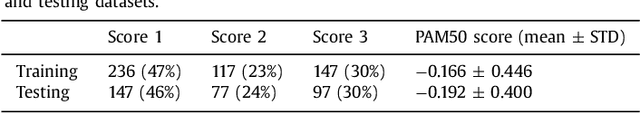
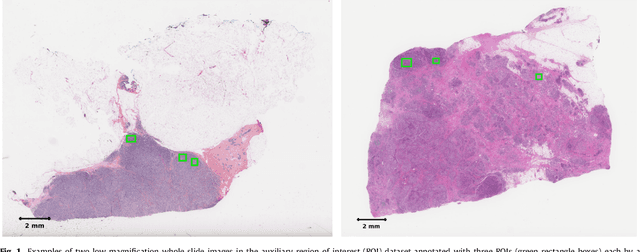
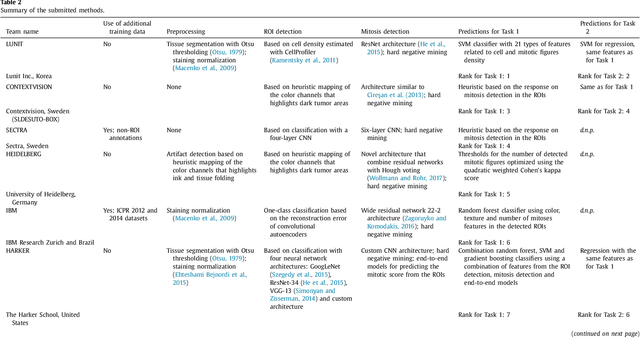
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
A Survey on Deep Learning in Medical Image Analysis
Jun 04, 2017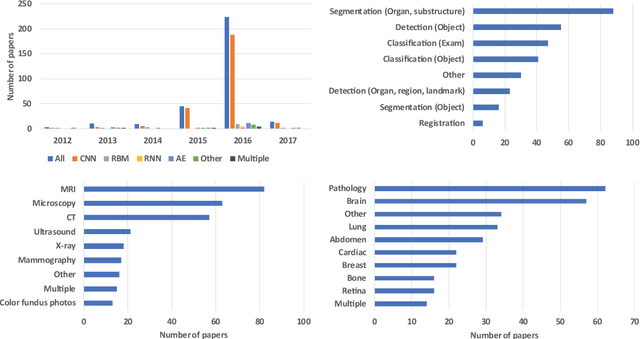
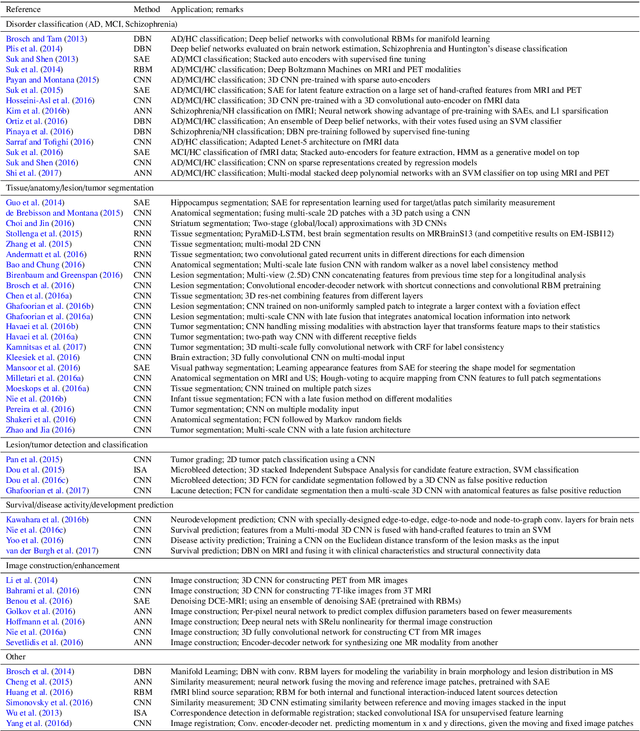
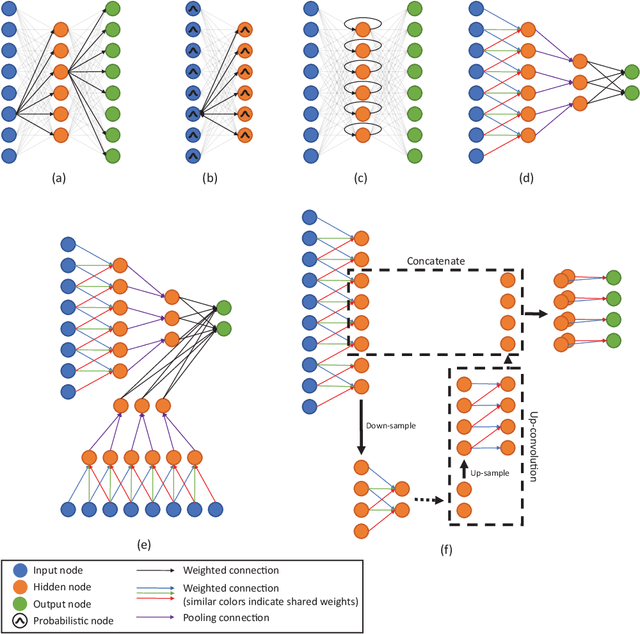
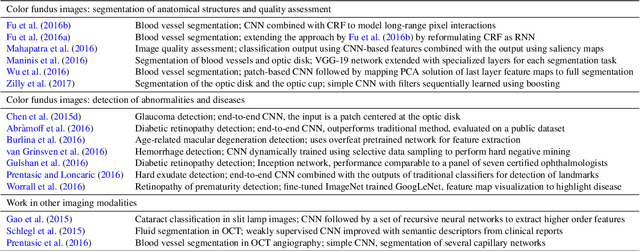
Abstract:Deep learning algorithms, in particular convolutional networks, have rapidly become a methodology of choice for analyzing medical images. This paper reviews the major deep learning concepts pertinent to medical image analysis and summarizes over 300 contributions to the field, most of which appeared in the last year. We survey the use of deep learning for image classification, object detection, segmentation, registration, and other tasks and provide concise overviews of studies per application area. Open challenges and directions for future research are discussed.
The importance of stain normalization in colorectal tissue classification with convolutional networks
May 23, 2017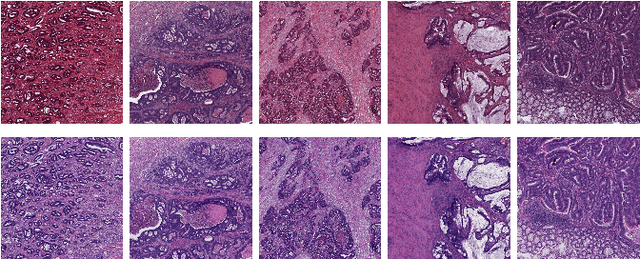

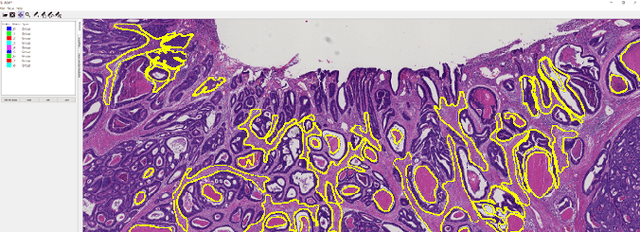
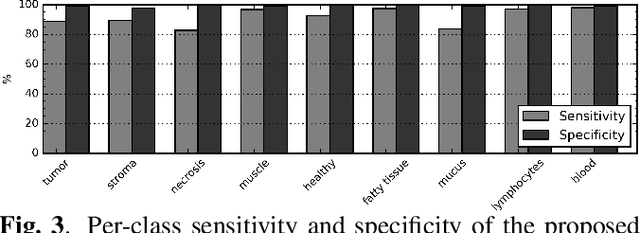
Abstract:The development of reliable imaging biomarkers for the analysis of colorectal cancer (CRC) in hematoxylin and eosin (H&E) stained histopathology images requires an accurate and reproducible classification of the main tissue components in the image. In this paper, we propose a system for CRC tissue classification based on convolutional networks (ConvNets). We investigate the importance of stain normalization in tissue classification of CRC tissue samples in H&E-stained images. Furthermore, we report the performance of ConvNets on a cohort of rectal cancer samples and on an independent publicly available dataset of colorectal H&E images.
Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images
May 10, 2017Abstract:Automated classification of histopathological whole-slide images (WSI) of breast tissue requires analysis at very high resolutions with a large contextual area. In this paper, we present context-aware stacked convolutional neural networks (CNN) for classification of breast WSIs into normal/benign, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC). We first train a CNN using high pixel resolution patches to capture cellular level information. The feature responses generated by this model are then fed as input to a second CNN, stacked on top of the first. Training of this stacked architecture with large input patches enables learning of fine-grained (cellular) details and global interdependence of tissue structures. Our system is trained and evaluated on a dataset containing 221 WSIs of H&E stained breast tissue specimens. The system achieves an AUC of 0.962 for the binary classification of non-malignant and malignant slides and obtains a three class accuracy of 81.3% for classification of WSIs into normal/benign, DCIS, and IDC, demonstrating its potentials for routine diagnostics.
Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images
Feb 19, 2017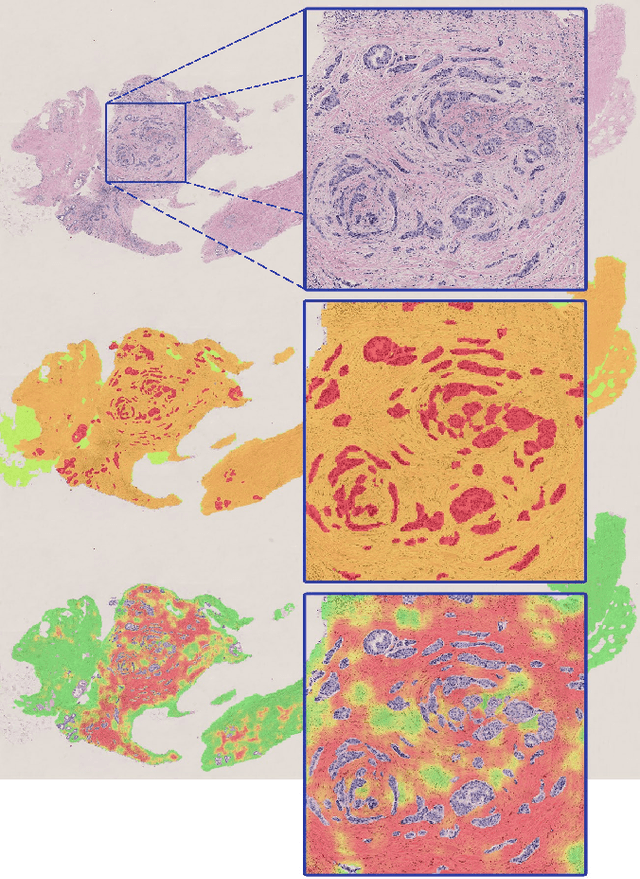
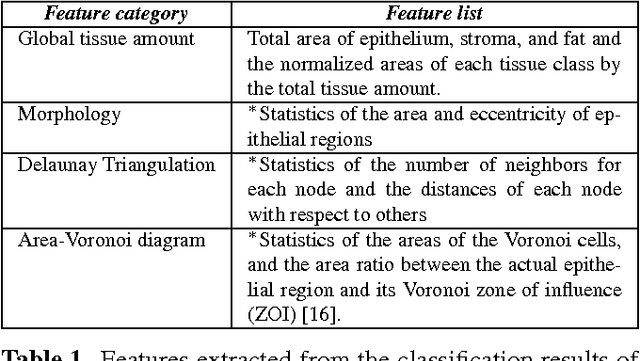
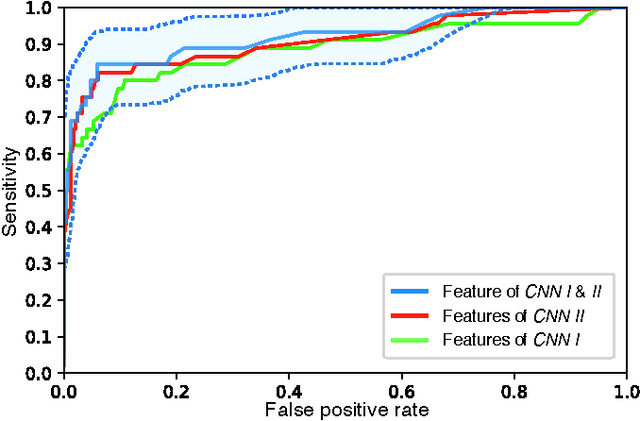
Abstract:Diagnosis of breast carcinomas has so far been limited to the morphological interpretation of epithelial cells and the assessment of epithelial tissue architecture. Consequently, most of the automated systems have focused on characterizing the epithelial regions of the breast to detect cancer. In this paper, we propose a system for classification of hematoxylin and eosin (H&E) stained breast specimens based on convolutional neural networks that primarily targets the assessment of tumor-associated stroma to diagnose breast cancer patients. We evaluate the performance of our proposed system using a large cohort containing 646 breast tissue biopsies. Our evaluations show that the proposed system achieves an area under ROC of 0.92, demonstrating the discriminative power of previously neglected tumor-associated stroma as a diagnostic biomarker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge