Alexandros Karargyris
IHU Strasbourg, UNISTRA
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022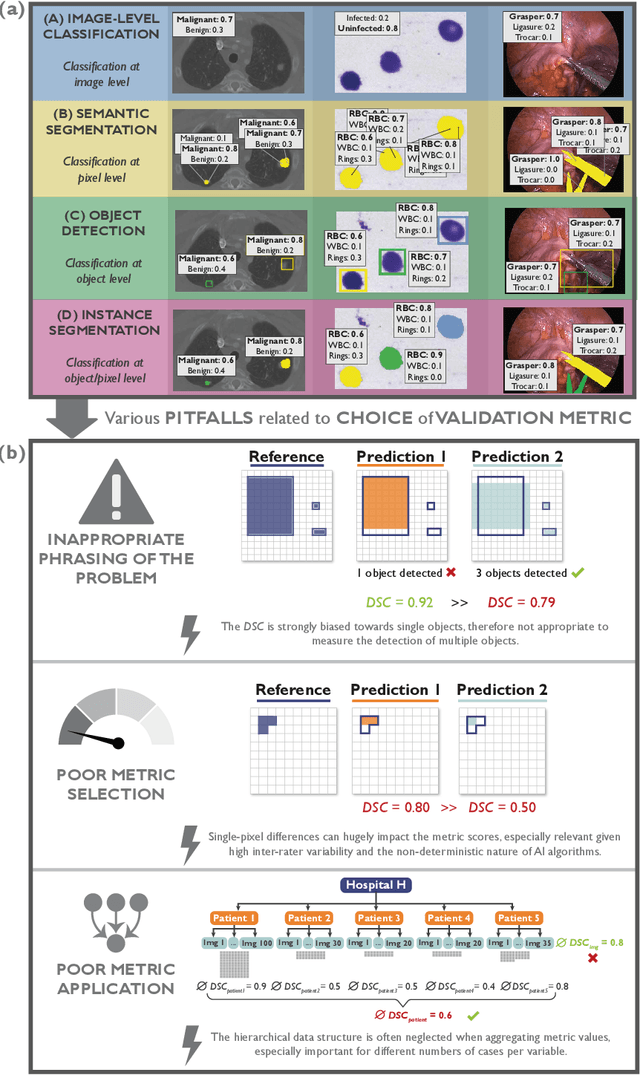
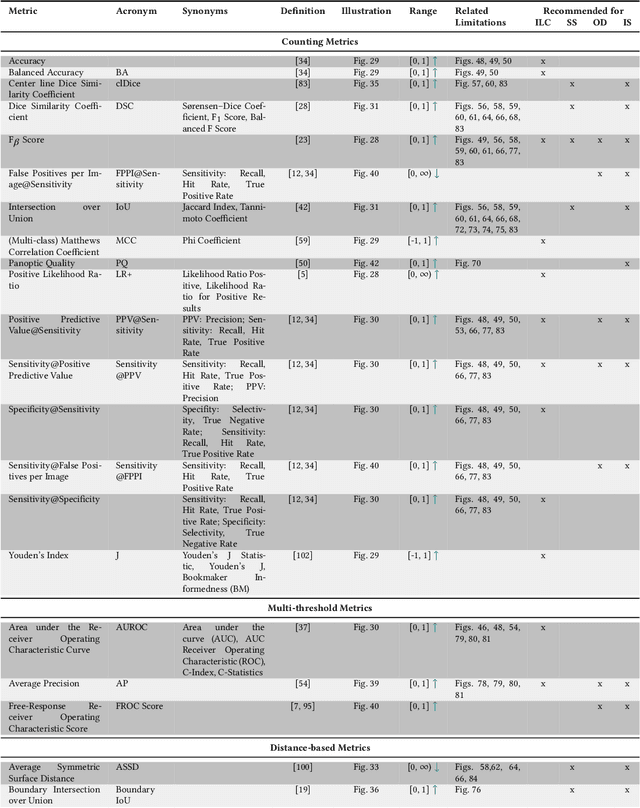
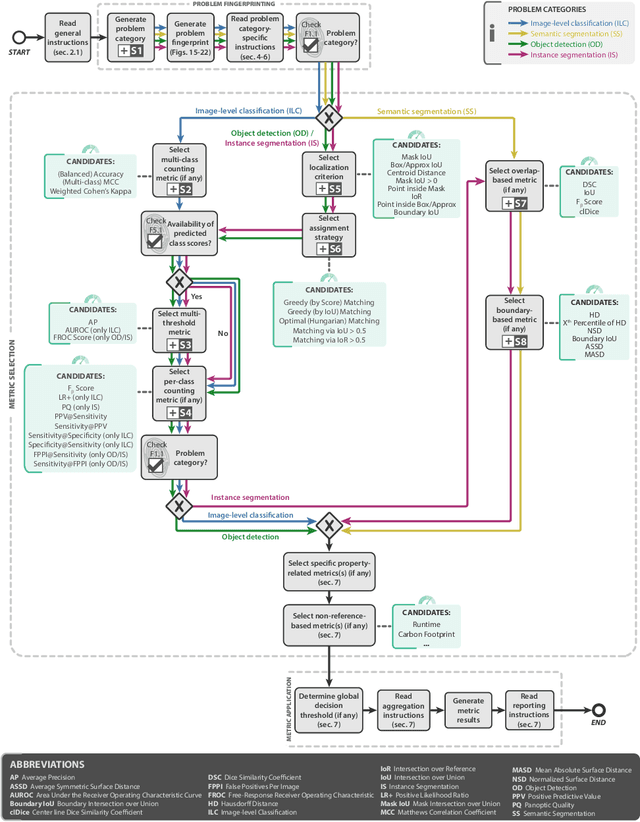
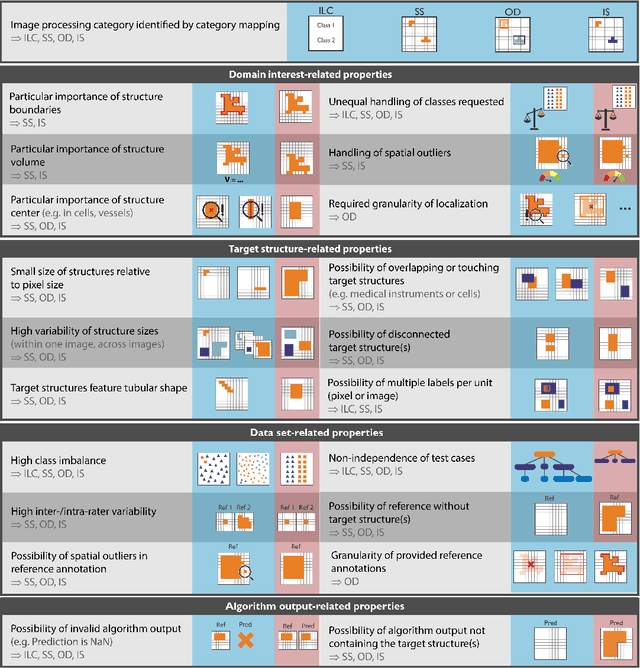
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
Federated Cycling (FedCy): Semi-supervised Federated Learning of Surgical Phases
Mar 14, 2022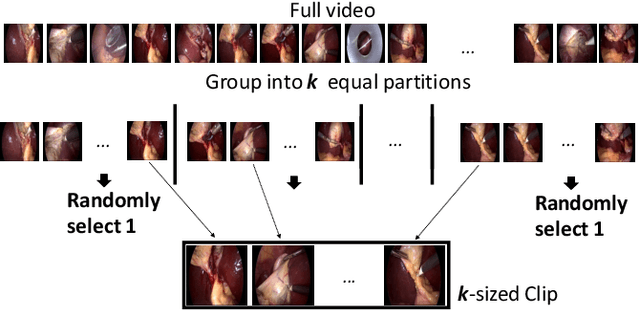
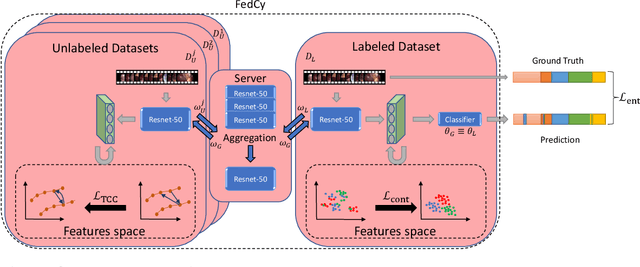
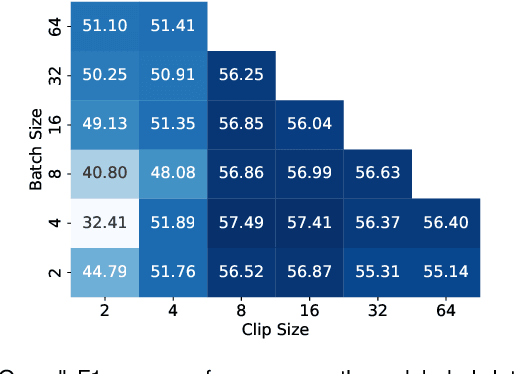
Abstract:Recent advancements in deep learning methods bring computer-assistance a step closer to fulfilling promises of safer surgical procedures. However, the generalizability of such methods is often dependent on training on diverse datasets from multiple medical institutions, which is a restrictive requirement considering the sensitive nature of medical data. Recently proposed collaborative learning methods such as Federated Learning (FL) allow for training on remote datasets without the need to explicitly share data. Even so, data annotation still represents a bottleneck, particularly in medicine and surgery where clinical expertise is often required. With these constraints in mind, we propose FedCy, a federated semi-supervised learning (FSSL) method that combines FL and self-supervised learning to exploit a decentralized dataset of both labeled and unlabeled videos, thereby improving performance on the task of surgical phase recognition. By leveraging temporal patterns in the labeled data, FedCy helps guide unsupervised training on unlabeled data towards learning task-specific features for phase recognition. We demonstrate significant performance gains over state-of-the-art FSSL methods on the task of automatic recognition of surgical phases using a newly collected multi-institutional dataset of laparoscopic cholecystectomy videos. Furthermore, we demonstrate that our approach also learns more generalizable features when tested on data from an unseen domain.
MedPerf: Open Benchmarking Platform for Medical Artificial Intelligence using Federated Evaluation
Oct 08, 2021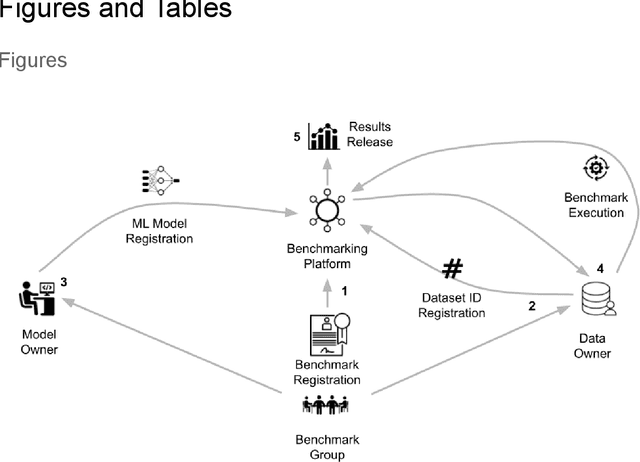
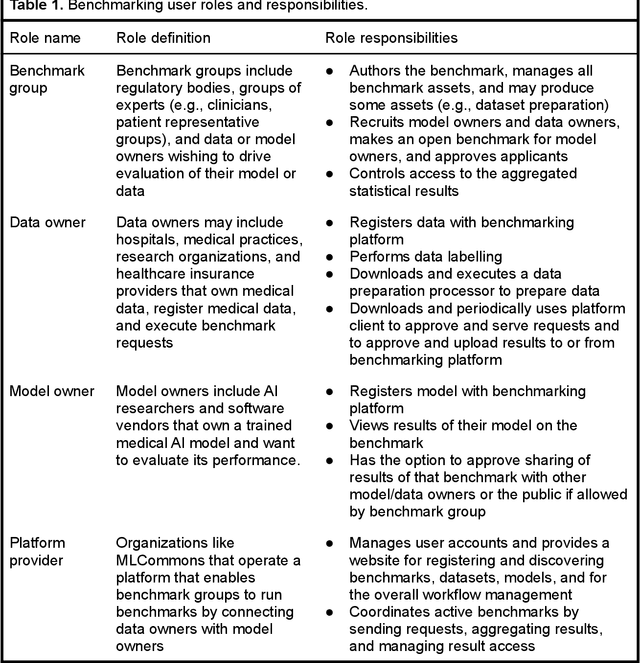
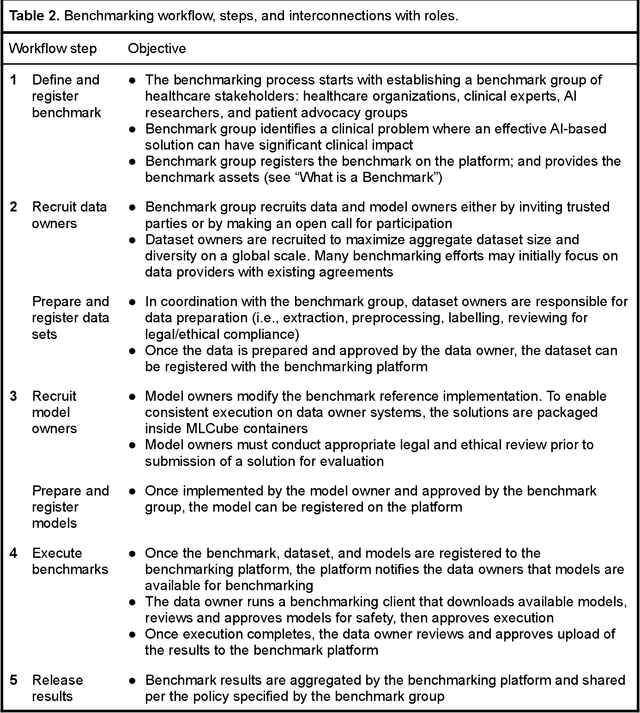
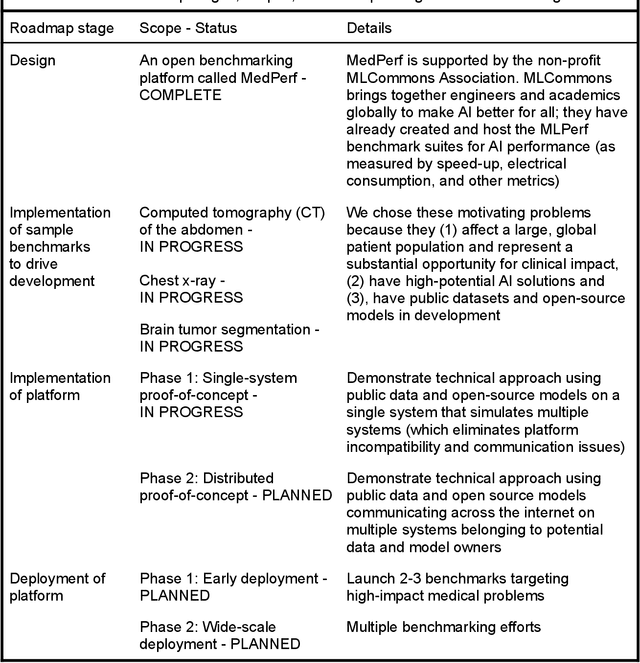
Abstract:Medical AI has tremendous potential to advance healthcare by supporting the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving provider and patient experience. We argue that unlocking this potential requires a systematic way to measure the performance of medical AI models on large-scale heterogeneous data. To meet this need, we are building MedPerf, an open framework for benchmarking machine learning in the medical domain. MedPerf will enable federated evaluation in which models are securely distributed to different facilities for evaluation, thereby empowering healthcare organizations to assess and verify the performance of AI models in an efficient and human-supervised process, while prioritizing privacy. We describe the current challenges healthcare and AI communities face, the need for an open platform, the design philosophy of MedPerf, its current implementation status, and our roadmap. We call for researchers and organizations to join us in creating the MedPerf open benchmarking platform.
Creation and Validation of a Chest X-Ray Dataset with Eye-tracking and Report Dictation for AI Development
Oct 08, 2020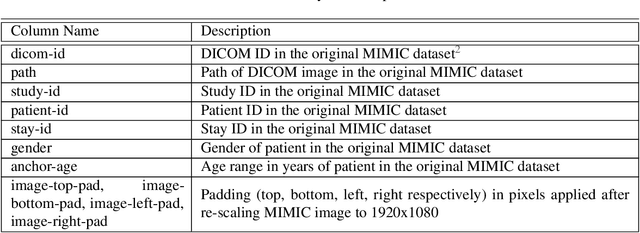
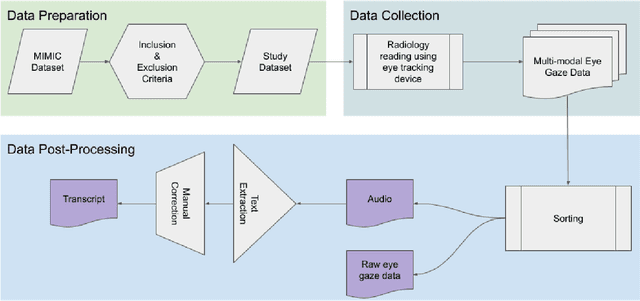
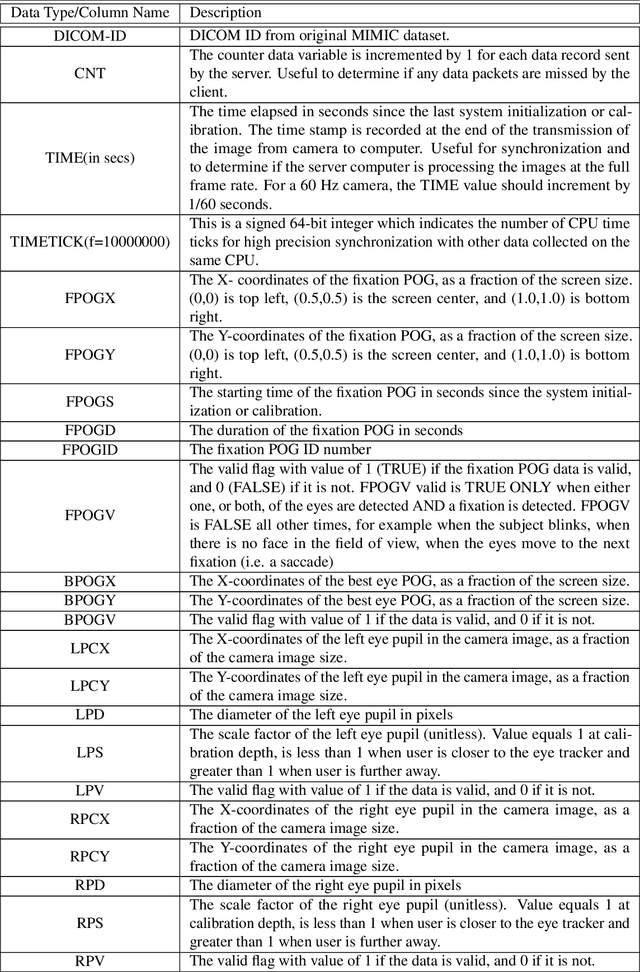
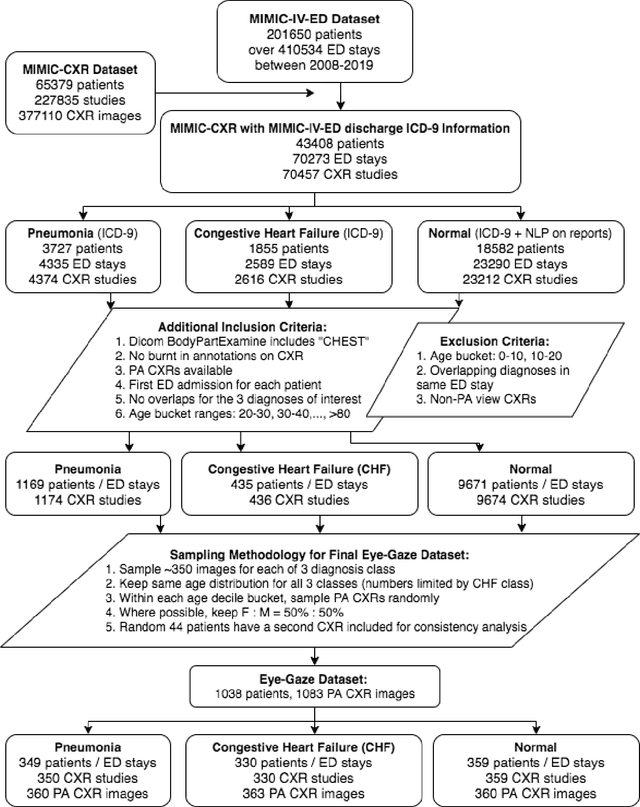
Abstract:We developed a rich dataset of Chest X-Ray (CXR) images to assist investigators in artificial intelligence. The data were collected using an eye tracking system while a radiologist reviewed and reported on 1,083 CXR images. The dataset contains the following aligned data: CXR image, transcribed radiology report text, radiologist's dictation audio and eye gaze coordinates data. We hope this dataset can contribute to various areas of research particularly towards explainable and multimodal deep learning / machine learning methods. Furthermore, investigators in disease classification and localization, automated radiology report generation, and human-machine interaction can benefit from these data. We report deep learning experiments that utilize the attention maps produced by eye gaze dataset to show the potential utility of this data.
Learning Invariant Feature Representation to Improve Generalization across Chest X-ray Datasets
Aug 04, 2020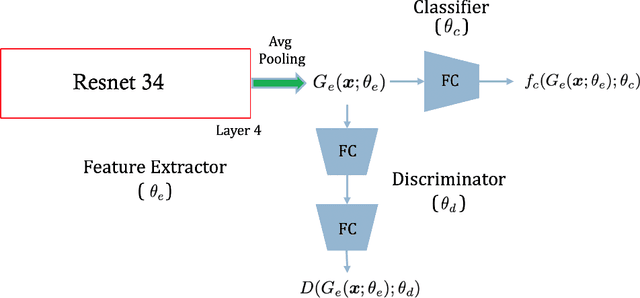
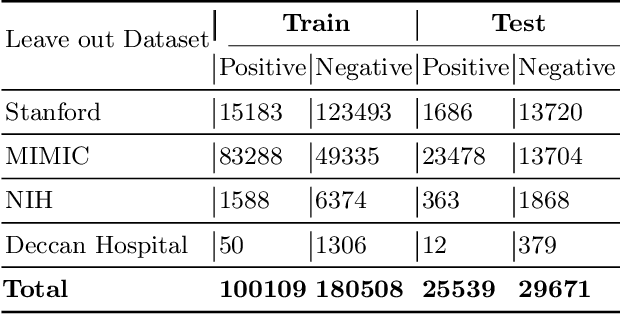
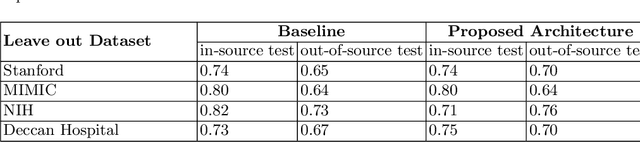
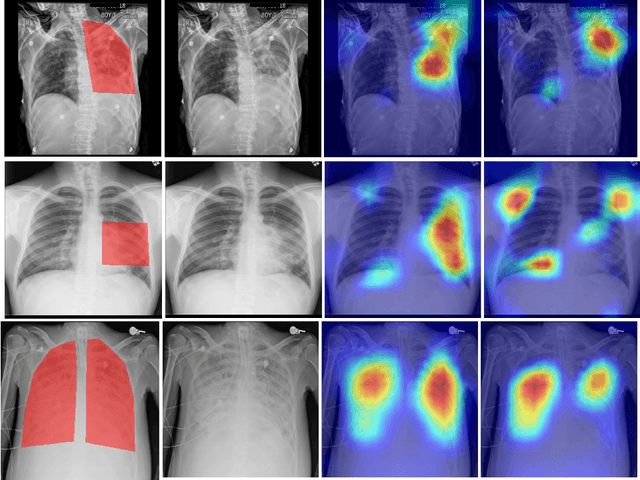
Abstract:Chest radiography is the most common medical image examination for screening and diagnosis in hospitals. Automatic interpretation of chest X-rays at the level of an entry-level radiologist can greatly benefit work prioritization and assist in analyzing a larger population. Subsequently, several datasets and deep learning-based solutions have been proposed to identify diseases based on chest X-ray images. However, these methods are shown to be vulnerable to shift in the source of data: a deep learning model performing well when tested on the same dataset as training data, starts to perform poorly when it is tested on a dataset from a different source. In this work, we address this challenge of generalization to a new source by forcing the network to learn a source-invariant representation. By employing an adversarial training strategy, we show that a network can be forced to learn a source-invariant representation. Through pneumonia-classification experiments on multi-source chest X-ray datasets, we show that this algorithm helps in improving classification accuracy on a new source of X-ray dataset.
Looking in the Right place for Anomalies: Explainable AI through Automatic Location Learning
Aug 02, 2020
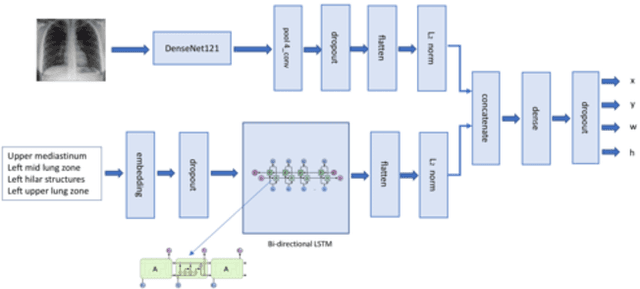
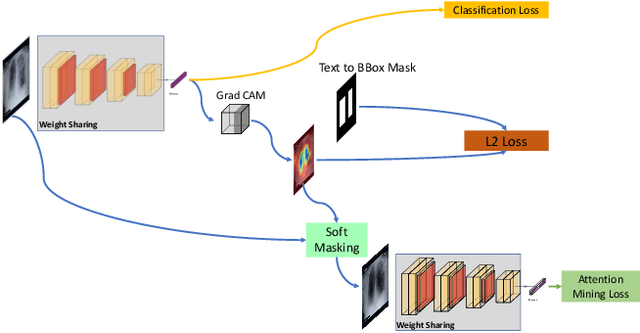
Abstract:Deep learning has now become the de facto approach to the recognition of anomalies in medical imaging. Their 'black box' way of classifying medical images into anomaly labels poses problems for their acceptance, particularly with clinicians. Current explainable AI methods offer justifications through visualizations such as heat maps but cannot guarantee that the network is focusing on the relevant image region fully containing the anomaly. In this paper, we develop an approach to explainable AI in which the anomaly is assured to be overlapping the expected location when present. This is made possible by automatically extracting location-specific labels from textual reports and learning the association of expected locations to labels using a hybrid combination of Bi-Directional Long Short-Term Memory Recurrent Neural Networks (Bi-LSTM) and DenseNet-121. Use of this expected location to bias the subsequent attention-guided inference network based on ResNet101 results in the isolation of the anomaly at the expected location when present. The method is evaluated on a large chest X-ray dataset.
* 5 pages, Paper presented as a poster at the International Symposium on Biomedical Imaging, 2020, Paper Number 655
Chest X-ray Report Generation through Fine-Grained Label Learning
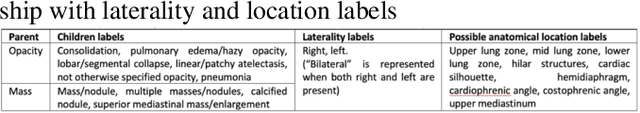
Jul 27, 2020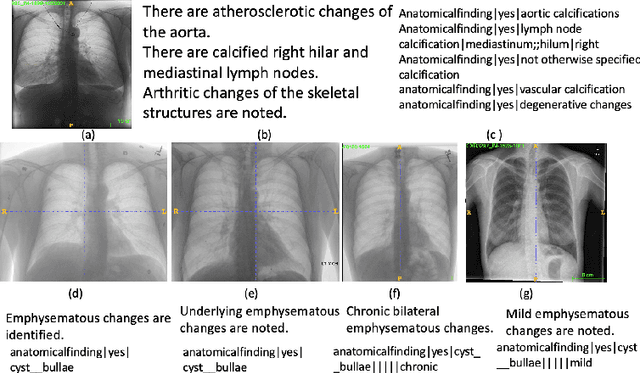
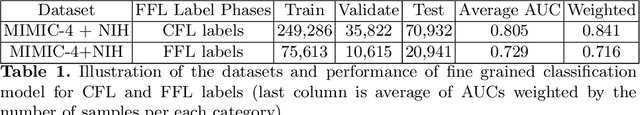
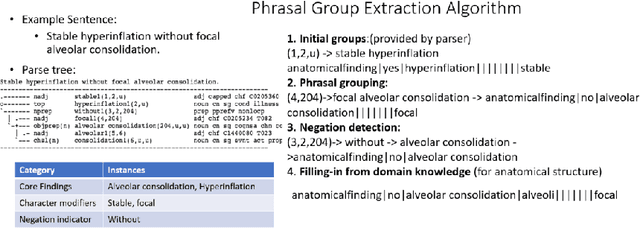
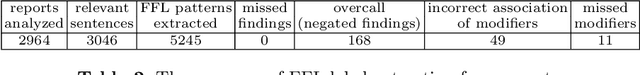
Abstract:Obtaining automated preliminary read reports for common exams such as chest X-rays will expedite clinical workflows and improve operational efficiencies in hospitals. However, the quality of reports generated by current automated approaches is not yet clinically acceptable as they cannot ensure the correct detection of a broad spectrum of radiographic findings nor describe them accurately in terms of laterality, anatomical location, severity, etc. In this work, we present a domain-aware automatic chest X-ray radiology report generation algorithm that learns fine-grained description of findings from images and uses their pattern of occurrences to retrieve and customize similar reports from a large report database. We also develop an automatic labeling algorithm for assigning such descriptors to images and build a novel deep learning network that recognizes both coarse and fine-grained descriptions of findings. The resulting report generation algorithm significantly outperforms the state of the art using established score metrics.
Self-Training with Improved Regularization for Few-Shot Chest X-Ray Classification
May 03, 2020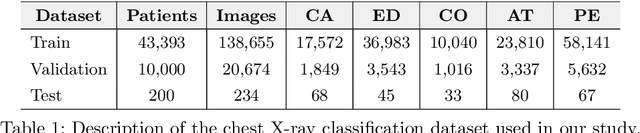
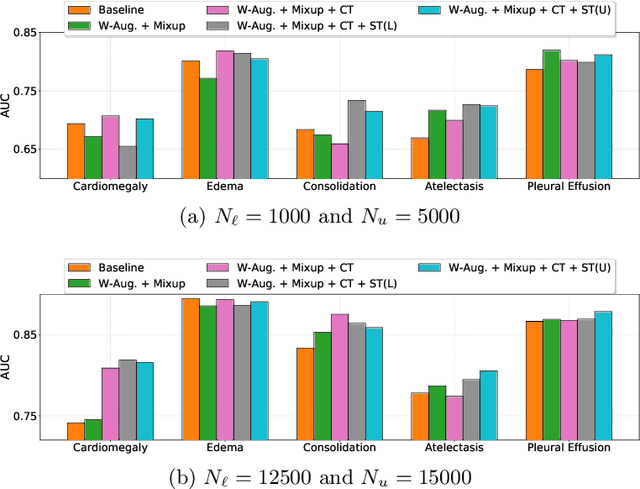
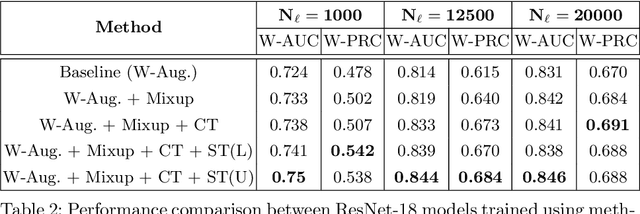
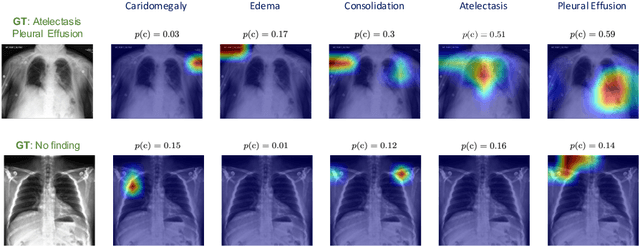
Abstract:Automated diagnostic assistants in healthcare necessitate accurate AI models that can be trained with limited labeled data, can cope with severe class imbalances and can support simultaneous prediction of multiple disease conditions. To this end, we present a novel few-shot learning approach that utilizes a number of key components to enable robust modeling in such challenging scenarios. Using an important use-case in chest X-ray classification, we provide several key insights on the effective use of data augmentation, self-training via distillation and confidence tempering for few-shot learning in medical imaging. Our results show that using only ~10% of the labeled data, we can build predictive models that match the performance of classifiers trained in a large-scale data setting.
Boosting the rule-out accuracy of deep disease detection using class weight modifiers
Jun 21, 2019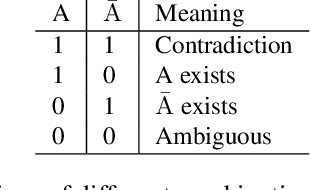
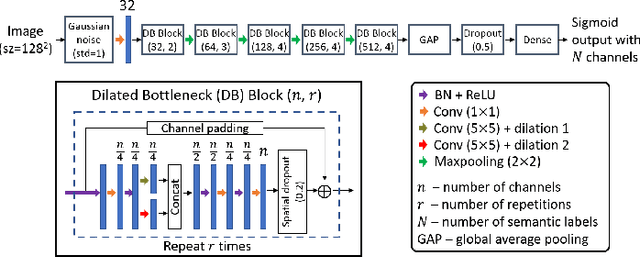
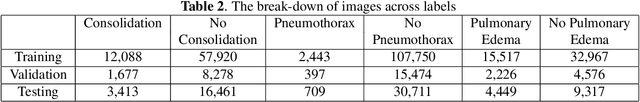
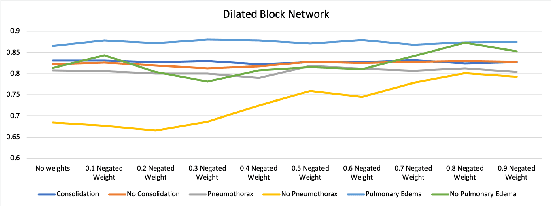
Abstract:In many screening applications, the primary goal of a radiologist or assisting artificial intelligence is to rule out certain findings. The classifiers built for such applications are often trained on large datasets that derive labels from clinical notes written for patients. While the quality of the positive findings described in these notes is often reliable, lack of the mention of a finding does not always rule out the presence of it. This happens because radiologists comment on the patient in the context of the exam, for example focusing on trauma as opposed to chronic disease at emergency rooms. However, this disease finding ambiguity can affect the performance of algorithms. Hence it is critical to model the ambiguity during training. We propose a scheme to apply reasonable class weight modifiers to our loss function for the no mention cases during training. We experiment with two different deep neural network architectures and show that the proposed method results in a large improvement in the performance of the classifiers, specially on negated findings. The baseline performance of a custom-made dilated block network proposed in this paper shows an improvement in comparison with baseline DenseNet-201, while both architectures benefit from the new proposed loss function weighting scheme. Over 200,000 chest X-ray images and three highly common diseases, along with their negated counterparts, are included in this study.
Building a Benchmark Dataset and Classifiers for Sentence-Level Findings in AP Chest X-rays
Jun 21, 2019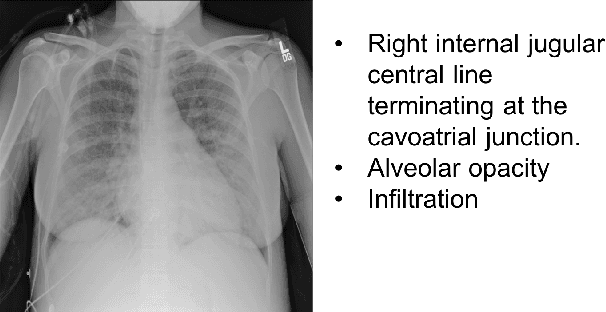
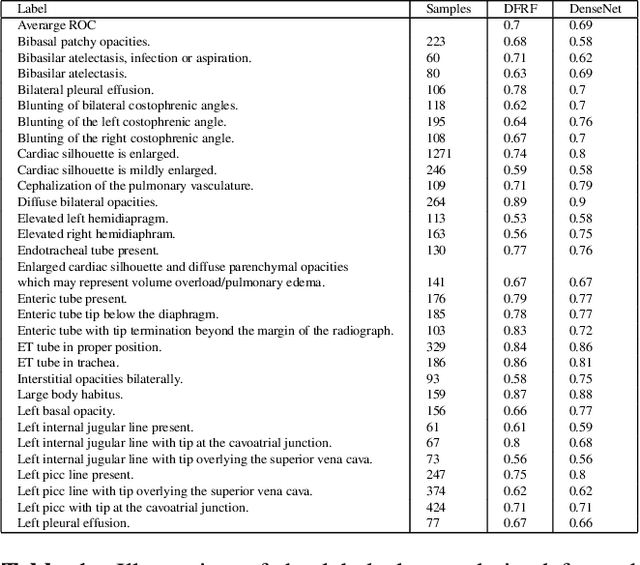
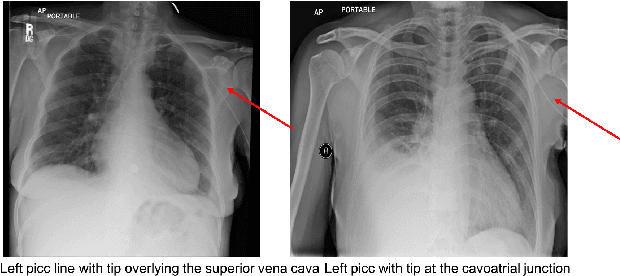
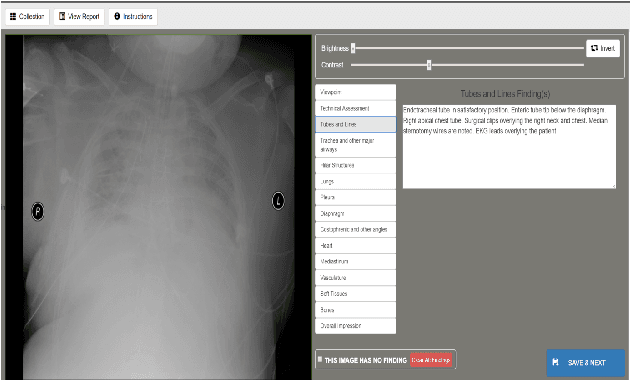
Abstract:Chest X-rays are the most common diagnostic exams in emergency rooms and hospitals. There has been a surge of work on automatic interpretation of chest X-rays using deep learning approaches after the availability of large open source chest X-ray dataset from NIH. However, the labels are not sufficiently rich and descriptive for training classification tools. Further, it does not adequately address the findings seen in Chest X-rays taken in anterior-posterior (AP) view which also depict the placement of devices such as central vascular lines and tubes. In this paper, we present a new chest X-ray benchmark database of 73 rich sentence-level descriptors of findings seen in AP chest X-rays. We describe our method of obtaining these findings through a semi-automated ground truth generation process from crowdsourcing of clinician annotations. We also present results of building classifiers for these findings that show that such higher granularity labels can also be learned through the framework of deep learning classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge