cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Advanced Deep Learning Techniques for Accurate Lung Cancer Detection and Classification
Aug 08, 2025Lung cancer (LC) ranks among the most frequently diagnosed cancers and is one of the most common causes of death for men and women worldwide. Computed Tomography (CT) images are the most preferred diagnosis method because of their low cost and their faster processing times. Many researchers have proposed various ways of identifying lung cancer using CT images. However, such techniques suffer from significant false positives, leading to low accuracy. The fundamental reason results from employing a small and imbalanced dataset. This paper introduces an innovative approach for LC detection and classification from CT images based on the DenseNet201 model. Our approach comprises several advanced methods such as Focal Loss, data augmentation, and regularization to overcome the imbalanced data issue and overfitting challenge. The findings show the appropriateness of the proposal, attaining a promising performance of 98.95% accuracy.
Transformer-Based Explainable Deep Learning for Breast Cancer Detection in Mammography: The MammoFormer Framework
Aug 08, 2025Breast cancer detection through mammography interpretation remains difficult because of the minimal nature of abnormalities that experts need to identify alongside the variable interpretations between readers. The potential of CNNs for medical image analysis faces two limitations: they fail to process both local information and wide contextual data adequately, and do not provide explainable AI (XAI) operations that doctors need to accept them in clinics. The researcher developed the MammoFormer framework, which unites transformer-based architecture with multi-feature enhancement components and XAI functionalities within one framework. Seven different architectures consisting of CNNs, Vision Transformer, Swin Transformer, and ConvNext were tested alongside four enhancement techniques, including original images, negative transformation, adaptive histogram equalization, and histogram of oriented gradients. The MammoFormer framework addresses critical clinical adoption barriers of AI mammography systems through: (1) systematic optimization of transformer architectures via architecture-specific feature enhancement, achieving up to 13% performance improvement, (2) comprehensive explainable AI integration providing multi-perspective diagnostic interpretability, and (3) a clinically deployable ensemble system combining CNN reliability with transformer global context modeling. The combination of transformer models with suitable feature enhancements enables them to achieve equal or better results than CNN approaches. ViT achieves 98.3% accuracy alongside AHE while Swin Transformer gains a 13.0% advantage through HOG enhancements
OCELOT 2023: Cell Detection from Cell-Tissue Interaction Challenge
Sep 11, 2025Pathologists routinely alternate between different magnifications when examining Whole-Slide Images, allowing them to evaluate both broad tissue morphology and intricate cellular details to form comprehensive diagnoses. However, existing deep learning-based cell detection models struggle to replicate these behaviors and learn the interdependent semantics between structures at different magnifications. A key barrier in the field is the lack of datasets with multi-scale overlapping cell and tissue annotations. The OCELOT 2023 challenge was initiated to gather insights from the community to validate the hypothesis that understanding cell and tissue (cell-tissue) interactions is crucial for achieving human-level performance, and to accelerate the research in this field. The challenge dataset includes overlapping cell detection and tissue segmentation annotations from six organs, comprising 673 pairs sourced from 306 The Cancer Genome Atlas (TCGA) Whole-Slide Images with hematoxylin and eosin staining, divided into training, validation, and test subsets. Participants presented models that significantly enhanced the understanding of cell-tissue relationships. Top entries achieved up to a 7.99 increase in F1-score on the test set compared to the baseline cell-only model that did not incorporate cell-tissue relationships. This is a substantial improvement in performance over traditional cell-only detection methods, demonstrating the need for incorporating multi-scale semantics into the models. This paper provides a comparative analysis of the methods used by participants, highlighting innovative strategies implemented in the OCELOT 2023 challenge.
* This is the accepted manuscript of an article published in Medical Image Analysis (Elsevier). The final version is available at: https://doi.org/10.1016/j.media.2025.103751
Optimizing Federated Learning Configurations for MRI Prostate Segmentation and Cancer Detection: A Simulation Study
Jul 30, 2025Purpose: To develop and optimize a federated learning (FL) framework across multiple clients for biparametric MRI prostate segmentation and clinically significant prostate cancer (csPCa) detection. Materials and Methods: A retrospective study was conducted using Flower FL to train a nnU-Net-based architecture for MRI prostate segmentation and csPCa detection, using data collected from January 2010 to August 2021. Model development included training and optimizing local epochs, federated rounds, and aggregation strategies for FL-based prostate segmentation on T2-weighted MRIs (four clients, 1294 patients) and csPCa detection using biparametric MRIs (three clients, 1440 patients). Performance was evaluated on independent test sets using the Dice score for segmentation and the Prostate Imaging: Cancer Artificial Intelligence (PI-CAI) score, defined as the average of the area under the receiver operating characteristic curve and average precision, for csPCa detection. P-values for performance differences were calculated using permutation testing. Results: The FL configurations were independently optimized for both tasks, showing improved performance at 1 epoch 300 rounds using FedMedian for prostate segmentation and 5 epochs 200 rounds using FedAdagrad, for csPCa detection. Compared with the average performance of the clients, the optimized FL model significantly improved performance in prostate segmentation and csPCa detection on the independent test set. The optimized FL model showed higher lesion detection performance compared to the FL-baseline model, but no evidence of a difference was observed for prostate segmentation. Conclusions: FL enhanced the performance and generalizability of MRI prostate segmentation and csPCa detection compared with local models, and optimizing its configuration further improved lesion detection performance.
Multi-Attention Stacked Ensemble for Lung Cancer Detection in CT Scans
Jul 27, 2025



In this work, we address the challenge of binary lung nodule classification (benign vs malignant) using CT images by proposing a multi-level attention stacked ensemble of deep neural networks. Three pretrained backbones - EfficientNet V2 S, MobileViT XXS, and DenseNet201 - are each adapted with a custom classification head tailored to 96 x 96 pixel inputs. A two-stage attention mechanism learns both model-wise and class-wise importance scores from concatenated logits, and a lightweight meta-learner refines the final prediction. To mitigate class imbalance and improve generalization, we employ dynamic focal loss with empirically calculated class weights, MixUp augmentation during training, and test-time augmentation at inference. Experiments on the LIDC-IDRI dataset demonstrate exceptional performance, achieving 98.09 accuracy and 0.9961 AUC, representing a 35 percent reduction in error rate compared to state-of-the-art methods. The model exhibits balanced performance across sensitivity (98.73) and specificity (98.96), with particularly strong results on challenging cases where radiologist disagreement was high. Statistical significance testing confirms the robustness of these improvements across multiple experimental runs. Our approach can serve as a robust, automated aid for radiologists in lung cancer screening.
Large Language Model Evaluated Stand-alone Attention-Assisted Graph Neural Network with Spatial and Structural Information Interaction for Precise Endoscopic Image Segmentation
Aug 09, 2025Accurate endoscopic image segmentation on the polyps is critical for early colorectal cancer detection. However, this task remains challenging due to low contrast with surrounding mucosa, specular highlights, and indistinct boundaries. To address these challenges, we propose FOCUS-Med, which stands for Fusion of spatial and structural graph with attentional context-aware polyp segmentation in endoscopic medical imaging. FOCUS-Med integrates a Dual Graph Convolutional Network (Dual-GCN) module to capture contextual spatial and topological structural dependencies. This graph-based representation enables the model to better distinguish polyps from background tissues by leveraging topological cues and spatial connectivity, which are often obscured in raw image intensities. It enhances the model's ability to preserve boundaries and delineate complex shapes typical of polyps. In addition, a location-fused stand-alone self-attention is employed to strengthen global context integration. To bridge the semantic gap between encoder-decoder layers, we incorporate a trainable weighted fast normalized fusion strategy for efficient multi-scale aggregation. Notably, we are the first to introduce the use of a Large Language Model (LLM) to provide detailed qualitative evaluations of segmentation quality. Extensive experiments on public benchmarks demonstrate that FOCUS-Med achieves state-of-the-art performance across five key metrics, underscoring its effectiveness and clinical potential for AI-assisted colonoscopy.
Enhanced Liver Tumor Detection in CT Images Using 3D U-Net and Bat Algorithm for Hyperparameter Optimization
Aug 11, 2025Liver cancer is one of the most prevalent and lethal forms of cancer, making early detection crucial for effective treatment. This paper introduces a novel approach for automated liver tumor segmentation in computed tomography (CT) images by integrating a 3D U-Net architecture with the Bat Algorithm for hyperparameter optimization. The method enhances segmentation accuracy and robustness by intelligently optimizing key parameters like the learning rate and batch size. Evaluated on a publicly available dataset, our model demonstrates a strong ability to balance precision and recall, with a high F1-score at lower prediction thresholds. This is particularly valuable for clinical diagnostics, where ensuring no potential tumors are missed is paramount. Our work contributes to the field of medical image analysis by demonstrating that the synergy between a robust deep learning architecture and a metaheuristic optimization algorithm can yield a highly effective solution for complex segmentation tasks.
Synthetic Data-Driven Multi-Architecture Framework for Automated Polyp Segmentation Through Integrated Detection and Mask Generation
Aug 08, 2025Colonoscopy is a vital tool for the early diagnosis of colorectal cancer, which is one of the main causes of cancer-related mortality globally; hence, it is deemed an essential technique for the prevention and early detection of colorectal cancer. The research introduces a unique multidirectional architectural framework to automate polyp detection within colonoscopy images while helping resolve limited healthcare dataset sizes and annotation complexities. The research implements a comprehensive system that delivers synthetic data generation through Stable Diffusion enhancements together with detection and segmentation algorithms. This detection approach combines Faster R-CNN for initial object localization while the Segment Anything Model (SAM) refines the segmentation masks. The faster R-CNN detection algorithm achieved a recall of 93.08% combined with a precision of 88.97% and an F1 score of 90.98%.SAM is then used to generate the image mask. The research evaluated five state-of-the-art segmentation models that included U-Net, PSPNet, FPN, LinkNet, and MANet using ResNet34 as a base model. The results demonstrate the superior performance of FPN with the highest scores of PSNR (7.205893) and SSIM (0.492381), while UNet excels in recall (84.85%) and LinkNet shows balanced performance in IoU (64.20%) and Dice score (77.53%).
Improving Diagnostic Accuracy for Oral Cancer with inpainting Synthesis Lesions Generated Using Diffusion Models
Aug 08, 2025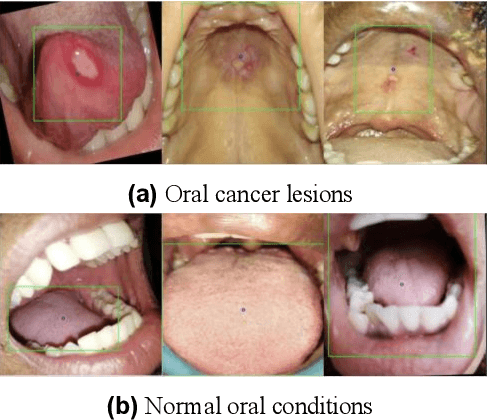
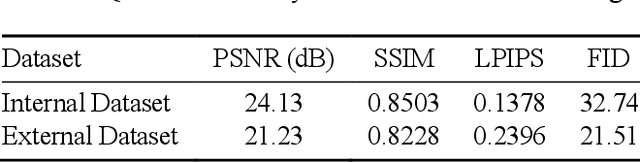
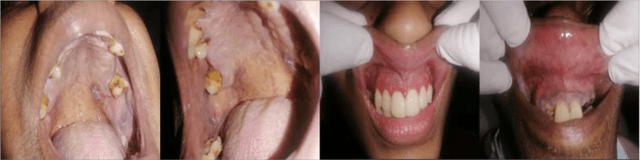
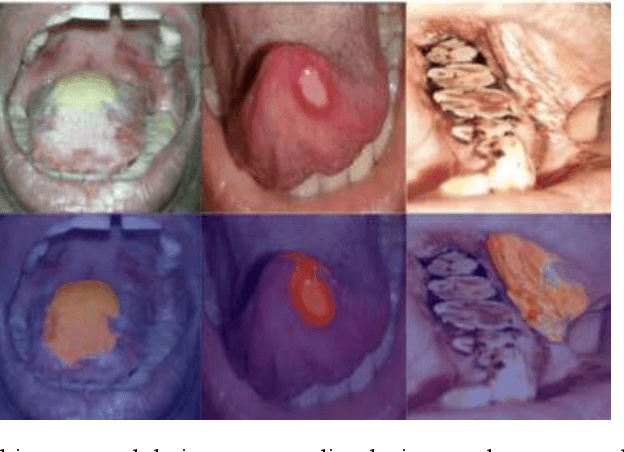
In oral cancer diagnostics, the limited availability of annotated datasets frequently constrains the performance of diagnostic models, particularly due to the variability and insufficiency of training data. To address these challenges, this study proposed a novel approach to enhance diagnostic accuracy by synthesizing realistic oral cancer lesions using an inpainting technique with a fine-tuned diffusion model. We compiled a comprehensive dataset from multiple sources, featuring a variety of oral cancer images. Our method generated synthetic lesions that exhibit a high degree of visual fidelity to actual lesions, thereby significantly enhancing the performance of diagnostic algorithms. The results show that our classification model achieved a diagnostic accuracy of 0.97 in differentiating between cancerous and non-cancerous tissues, while our detection model accurately identified lesion locations with 0.85 accuracy. This method validates the potential for synthetic image generation in medical diagnostics and paves the way for further research into extending these methods to other types of cancer diagnostics.
Model Compression Engine for Wearable Devices Skin Cancer Diagnosis
Jul 23, 2025Skin cancer is one of the most prevalent and preventable types of cancer, yet its early detection remains a challenge, particularly in resource-limited settings where access to specialized healthcare is scarce. This study proposes an AI-driven diagnostic tool optimized for embedded systems to address this gap. Using transfer learning with the MobileNetV2 architecture, the model was adapted for binary classification of skin lesions into "Skin Cancer" and "Other." The TensorRT framework was employed to compress and optimize the model for deployment on the NVIDIA Jetson Orin Nano, balancing performance with energy efficiency. Comprehensive evaluations were conducted across multiple benchmarks, including model size, inference speed, throughput, and power consumption. The optimized models maintained their performance, achieving an F1-Score of 87.18% with a precision of 93.18% and recall of 81.91%. Post-compression results showed reductions in model size of up to 0.41, along with improvements in inference speed and throughput, and a decrease in energy consumption of up to 0.93 in INT8 precision. These findings validate the feasibility of deploying high-performing, energy-efficient diagnostic tools on resource-constrained edge devices. Beyond skin cancer detection, the methodologies applied in this research have broader applications in other medical diagnostics and domains requiring accessible, efficient AI solutions. This study underscores the potential of optimized AI systems to revolutionize healthcare diagnostics, thereby bridging the divide between advanced technology and underserved regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge