cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
Artificial Intelligence-Enabled Analysis of Radiology Reports: Epidemiology and Consequences of Incidental Thyroid Findings
Oct 30, 2025



Importance Incidental thyroid findings (ITFs) are increasingly detected on imaging performed for non-thyroid indications. Their prevalence, features, and clinical consequences remain undefined. Objective To develop, validate, and deploy a natural language processing (NLP) pipeline to identify ITFs in radiology reports and assess their prevalence, features, and clinical outcomes. Design, Setting, and Participants Retrospective cohort of adults without prior thyroid disease undergoing thyroid-capturing imaging at Mayo Clinic sites from July 1, 2017, to September 30, 2023. A transformer-based NLP pipeline identified ITFs and extracted nodule characteristics from image reports from multiple modalities and body regions. Main Outcomes and Measures Prevalence of ITFs, downstream thyroid ultrasound, biopsy, thyroidectomy, and thyroid cancer diagnosis. Logistic regression identified demographic and imaging-related factors. Results Among 115,683 patients (mean age, 56.8 [SD 17.2] years; 52.9% women), 9,077 (7.8%) had an ITF, of which 92.9% were nodules. ITFs were more likely in women, older adults, those with higher BMI, and when imaging was ordered by oncology or internal medicine. Compared with chest CT, ITFs were more likely via neck CT, PET, and nuclear medicine scans. Nodule characteristics were poorly documented, with size reported in 44% and other features in fewer than 15% (e.g. calcifications). Compared with patients without ITFs, those with ITFs had higher odds of thyroid nodule diagnosis, biopsy, thyroidectomy and thyroid cancer diagnosis. Most cancers were papillary, and larger when detected after ITFs vs no ITF. Conclusions ITFs were common and strongly associated with cascades leading to the detection of small, low-risk cancers. These findings underscore the role of ITFs in thyroid cancer overdiagnosis and the need for standardized reporting and more selective follow-up.
Scale-Aware Curriculum Learning for Ddata-Efficient Lung Nodule Detection with YOLOv11
Oct 30, 2025Lung nodule detection in chest CT is crucial for early lung cancer diagnosis, yet existing deep learning approaches face challenges when deployed in clinical settings with limited annotated data. While curriculum learning has shown promise in improving model training, traditional static curriculum strategies fail in data-scarce scenarios. We propose Scale Adaptive Curriculum Learning (SACL), a novel training strategy that dynamically adjusts curriculum design based on available data scale. SACL introduces three key mechanisms:(1) adaptive epoch scheduling, (2) hard sample injection, and (3) scale-aware optimization. We evaluate SACL on the LUNA25 dataset using YOLOv11 as the base detector. Experimental results demonstrate that while SACL achieves comparable performance to static curriculum learning on the full dataset in mAP50, it shows significant advantages under data-limited conditions with 4.6%, 3.5%, and 2.0% improvements over baseline at 10%, 20%, and 50% of training data respectively. By enabling robust training across varying data scales without architectural modifications, SACL provides a practical solution for healthcare institutions to develop effective lung nodule detection systems despite limited annotation resources.
SENCA-st: Integrating Spatial Transcriptomics and Histopathology with Cross Attention Shared Encoder for Region Identification in Cancer Pathology
Nov 11, 2025Spatial transcriptomics is an emerging field that enables the identification of functional regions based on the spatial distribution of gene expression. Integrating this functional information present in transcriptomic data with structural data from histopathology images is an active research area with applications in identifying tumor substructures associated with cancer drug resistance. Current histopathology-spatial-transcriptomic region segmentation methods suffer due to either making spatial transcriptomics prominent by using histopathology features just to assist processing spatial transcriptomics data or using vanilla contrastive learning that make histopathology images prominent due to only promoting common features losing functional information. In both extremes, the model gets either lost in the noise of spatial transcriptomics or overly smoothed, losing essential information. Thus, we propose our novel architecture SENCA-st (Shared Encoder with Neighborhood Cross Attention) that preserves the features of both modalities. More importantly, it emphasizes regions that are structurally similar in histopathology but functionally different on spatial transcriptomics using cross-attention. We demonstrate the superior performance of our model that surpasses state-of-the-art methods in detecting tumor heterogeneity and tumor micro-environment regions, a clinically crucial aspect.
A Clinical-grade Universal Foundation Model for Intraoperative Pathology
Oct 06, 2025Intraoperative pathology is pivotal to precision surgery, yet its clinical impact is constrained by diagnostic complexity and the limited availability of high-quality frozen-section data. While computational pathology has made significant strides, the lack of large-scale, prospective validation has impeded its routine adoption in surgical workflows. Here, we introduce CRISP, a clinical-grade foundation model developed on over 100,000 frozen sections from eight medical centers, specifically designed to provide Clinical-grade Robust Intraoperative Support for Pathology (CRISP). CRISP was comprehensively evaluated on more than 15,000 intraoperative slides across nearly 100 retrospective diagnostic tasks, including benign-malignant discrimination, key intraoperative decision-making, and pan-cancer detection, etc. The model demonstrated robust generalization across diverse institutions, tumor types, and anatomical sites-including previously unseen sites and rare cancers. In a prospective cohort of over 2,000 patients, CRISP sustained high diagnostic accuracy under real-world conditions, directly informing surgical decisions in 92.6% of cases. Human-AI collaboration further reduced diagnostic workload by 35%, avoided 105 ancillary tests and enhanced detection of micrometastases with 87.5% accuracy. Together, these findings position CRISP as a clinical-grade paradigm for AI-driven intraoperative pathology, bridging computational advances with surgical precision and accelerating the translation of artificial intelligence into routine clinical practice.
Transformer Classification of Breast Lesions: The BreastDCEDL_AMBL Benchmark Dataset and 0.92 AUC Baseline
Sep 30, 2025



The error is caused by special characters that arXiv's system doesn't recognize. Here's the cleaned version with all problematic characters replaced: Breast magnetic resonance imaging is a critical tool for cancer detection and treatment planning, but its clinical utility is hindered by poor specificity, leading to high false-positive rates and unnecessary biopsies. This study introduces a transformer-based framework for automated classification of breast lesions in dynamic contrast-enhanced MRI, addressing the challenge of distinguishing benign from malignant findings. We implemented a SegFormer architecture that achieved an AUC of 0.92 for lesion-level classification, with 100% sensitivity and 67% specificity at the patient level - potentially eliminating one-third of unnecessary biopsies without missing malignancies. The model quantifies malignant pixel distribution via semantic segmentation, producing interpretable spatial predictions that support clinical decision-making. To establish reproducible benchmarks, we curated BreastDCEDL_AMBL by transforming The Cancer Imaging Archive's AMBL collection into a standardized deep learning dataset with 88 patients and 133 annotated lesions (89 benign, 44 malignant). This resource addresses a key infrastructure gap, as existing public datasets lack benign lesion annotations, limiting benign-malignant classification research. Training incorporated an expanded cohort of over 1,200 patients through integration with BreastDCEDL datasets, validating transfer learning approaches despite primary tumor-only annotations. Public release of the dataset, models, and evaluation protocols provides the first standardized benchmark for DCE-MRI lesion classification, enabling methodological advancement toward clinical deployment.
Breast Cancer Detection in Thermographic Images via Diffusion-Based Augmentation and Nonlinear Feature Fusion
Sep 08, 2025Data scarcity hinders deep learning for medical imaging. We propose a framework for breast cancer classification in thermograms that addresses this using a Diffusion Probabilistic Model (DPM) for data augmentation. Our DPM-based augmentation is shown to be superior to both traditional methods and a ProGAN baseline. The framework fuses deep features from a pre-trained ResNet-50 with handcrafted nonlinear features (e.g., Fractal Dimension) derived from U-Net segmented tumors. An XGBoost classifier trained on these fused features achieves 98.0\% accuracy and 98.1\% sensitivity. Ablation studies and statistical tests confirm that both the DPM augmentation and the nonlinear feature fusion are critical, statistically significant components of this success. This work validates the synergy between advanced generative models and interpretable features for creating highly accurate medical diagnostic tools.
Interpretable Deep Transfer Learning for Breast Ultrasound Cancer Detection: A Multi-Dataset Study
Sep 05, 2025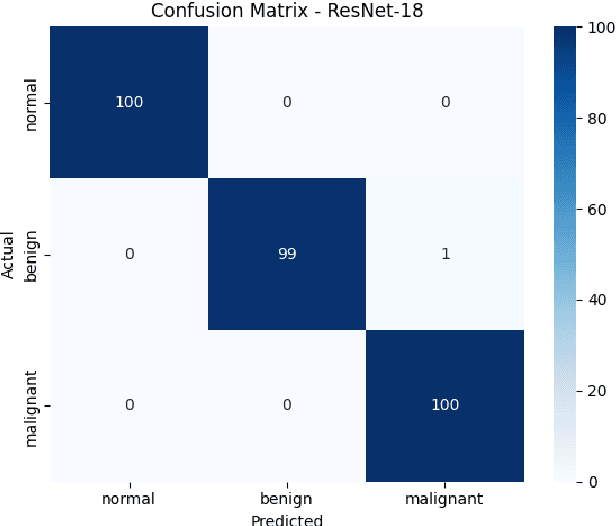
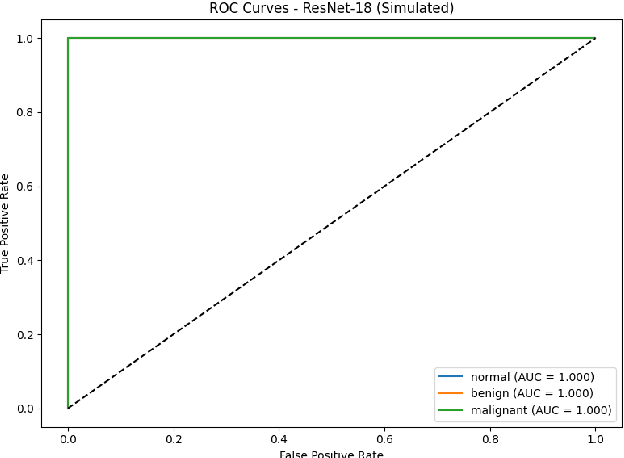
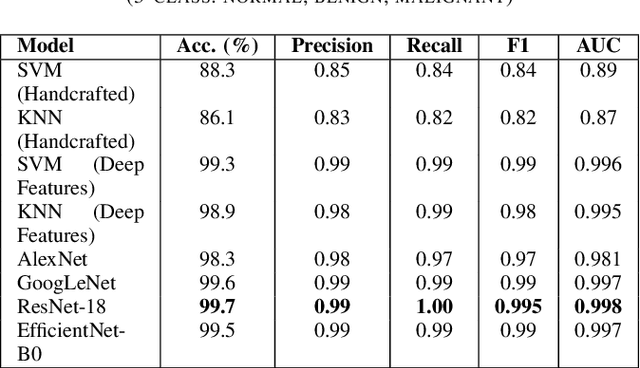
Breast cancer remains a leading cause of cancer-related mortality among women worldwide. Ultrasound imaging, widely used due to its safety and cost-effectiveness, plays a key role in early detection, especially in patients with dense breast tissue. This paper presents a comprehensive study on the application of machine learning and deep learning techniques for breast cancer classification using ultrasound images. Using datasets such as BUSI, BUS-BRA, and BrEaST-Lesions USG, we evaluate classical machine learning models (SVM, KNN) and deep convolutional neural networks (ResNet-18, EfficientNet-B0, GoogLeNet). Experimental results show that ResNet-18 achieves the highest accuracy (99.7%) and perfect sensitivity for malignant lesions. Classical ML models, though outperformed by CNNs, achieve competitive performance when enhanced with deep feature extraction. Grad-CAM visualizations further improve model transparency by highlighting diagnostically relevant image regions. These findings support the integration of AI-based diagnostic tools into clinical workflows and demonstrate the feasibility of deploying high-performing, interpretable systems for ultrasound-based breast cancer detection.
Prostate Capsule Segmentation from Micro-Ultrasound Images using Adaptive Focal Loss
Sep 19, 2025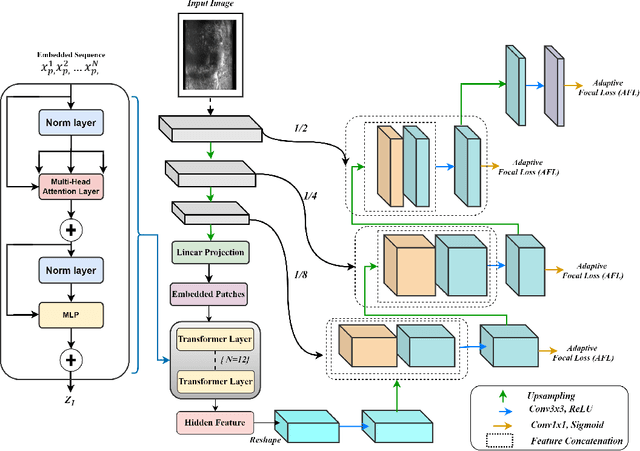
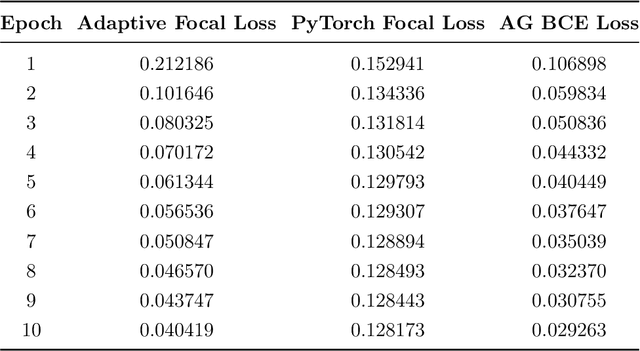
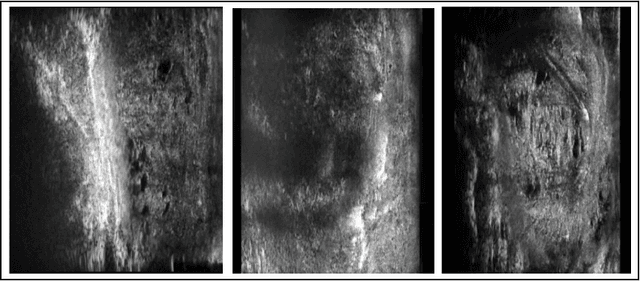
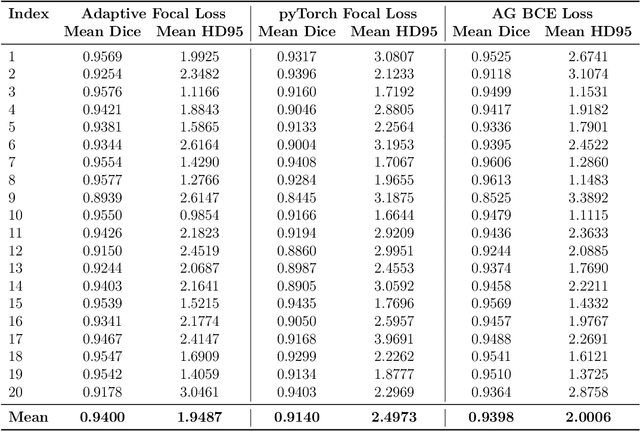
Micro-ultrasound (micro-US) is a promising imaging technique for cancer detection and computer-assisted visualization. This study investigates prostate capsule segmentation using deep learning techniques from micro-US images, addressing the challenges posed by the ambiguous boundaries of the prostate capsule. Existing methods often struggle in such cases, motivating the development of a tailored approach. This study introduces an adaptive focal loss function that dynamically emphasizes both hard and easy regions, taking into account their respective difficulty levels and annotation variability. The proposed methodology has two primary strategies: integrating a standard focal loss function as a baseline to design an adaptive focal loss function for proper prostate capsule segmentation. The focal loss baseline provides a robust foundation, incorporating class balancing and focusing on examples that are difficult to classify. The adaptive focal loss offers additional flexibility, addressing the fuzzy region of the prostate capsule and annotation variability by dilating the hard regions identified through discrepancies between expert and non-expert annotations. The proposed method dynamically adjusts the segmentation model's weights better to identify the fuzzy regions of the prostate capsule. The proposed adaptive focal loss function demonstrates superior performance, achieving a mean dice coefficient (DSC) of 0.940 and a mean Hausdorff distance (HD) of 1.949 mm in the testing dataset. These results highlight the effectiveness of integrating advanced loss functions and adaptive techniques into deep learning models. This enhances the accuracy of prostate capsule segmentation in micro-US images, offering the potential to improve clinical decision-making in prostate cancer diagnosis and treatment planning.
Segmentation and Classification of Pap Smear Images for Cervical Cancer Detection Using Deep Learning
Aug 25, 2025Cervical cancer remains a significant global health concern and a leading cause of cancer-related deaths among women. Early detection through Pap smear tests is essential to reduce mortality rates; however, the manual examination is time consuming and prone to human error. This study proposes a deep learning framework that integrates U-Net for segmentation and a classification model to enhance diagnostic performance. The Herlev Pap Smear Dataset, a publicly available cervical cell dataset, was utilized for training and evaluation. The impact of segmentation on classification performance was evaluated by comparing the model trained on segmented images and another trained on non-segmented images. Experimental results showed that the use of segmented images marginally improved the model performance on precision (about 0.41 percent higher) and F1-score (about 1.30 percent higher), which suggests a slightly more balanced classification performance. While segmentation helps in feature extraction, the results showed that its impact on classification performance appears to be limited. The proposed framework offers a supplemental tool for clinical applications, which may aid pathologists in early diagnosis.
Deep Learning Framework for Early Detection of Pancreatic Cancer Using Multi-Modal Medical Imaging Analysis
Aug 28, 2025Pacreatic ductal adenocarcinoma (PDAC) remains one of the most lethal forms of cancer, with a five-year survival rate below 10% primarily due to late detection. This research develops and validates a deep learning framework for early PDAC detection through analysis of dual-modality imaging: autofluorescence and second harmonic generation (SHG). We analyzed 40 unique patient samples to create a specialized neural network capable of distinguishing between normal, fibrotic, and cancerous tissue. Our methodology evaluated six distinct deep learning architectures, comparing traditional Convolutional Neural Networks (CNNs) with modern Vision Transformers (ViTs). Through systematic experimentation, we identified and overcome significant challenges in medical image analysis, including limited dataset size and class imbalance. The final optimized framework, based on a modified ResNet architecture with frozen pre-trained layers and class-weighted training, achieved over 90% accuracy in cancer detection. This represents a significant improvement over current manual analysis methods an demonstrates potential for clinical deployment. This work establishes a robust pipeline for automated PDAC detection that can augment pathologists' capabilities while providing a foundation for future expansion to other cancer types. The developed methodology also offers valuable insights for applying deep learning to limited-size medical imaging datasets, a common challenge in clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge