cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
DGSAN: Dual-Graph Spatiotemporal Attention Network for Pulmonary Nodule Malignancy Prediction
Dec 24, 2025Lung cancer continues to be the leading cause of cancer-related deaths globally. Early detection and diagnosis of pulmonary nodules are essential for improving patient survival rates. Although previous research has integrated multimodal and multi-temporal information, outperforming single modality and single time point, the fusion methods are limited to inefficient vector concatenation and simple mutual attention, highlighting the need for more effective multimodal information fusion. To address these challenges, we introduce a Dual-Graph Spatiotemporal Attention Network, which leverages temporal variations and multimodal data to enhance the accuracy of predictions. Our methodology involves developing a Global-Local Feature Encoder to better capture the local, global, and fused characteristics of pulmonary nodules. Additionally, a Dual-Graph Construction method organizes multimodal features into inter-modal and intra-modal graphs. Furthermore, a Hierarchical Cross-Modal Graph Fusion Module is introduced to refine feature integration. We also compiled a novel multimodal dataset named the NLST-cmst dataset as a comprehensive source of support for related research. Our extensive experiments, conducted on both the NLST-cmst and curated CSTL-derived datasets, demonstrate that our DGSAN significantly outperforms state-of-the-art methods in classifying pulmonary nodules with exceptional computational efficiency.
Lesion Segmentation in FDG-PET/CT Using Swin Transformer U-Net 3D: A Robust Deep Learning Framework
Jan 06, 2026Accurate and automated lesion segmentation in Positron Emission Tomography / Computed Tomography (PET/CT) imaging is essential for cancer diagnosis and therapy planning. This paper presents a Swin Transformer UNet 3D (SwinUNet3D) framework for lesion segmentation in Fluorodeoxyglucose Positron Emission Tomography / Computed Tomography (FDG-PET/CT) scans. By combining shifted window self-attention with U-Net style skip connections, the model captures both global context and fine anatomical detail. We evaluate SwinUNet3D on the AutoPET III FDG dataset and compare it against a baseline 3D U-Net. Results show that SwinUNet3D achieves a Dice score of 0.88 and IoU of 0.78, surpassing 3D U-Net (Dice 0.48, IoU 0.32) while also delivering faster inference times. Qualitative analysis demonstrates improved detection of small and irregular lesions, reduced false positives, and more accurate PET/CT fusion. While the framework is currently limited to FDG scans and trained under modest GPU resources, it establishes a strong foundation for future multi-tracer, multi-center evaluations and benchmarking against other transformer-based architectures. Overall, SwinUNet3D represents an efficient and robust approach to PET/CT lesion segmentation, advancing the integration of transformer-based models into oncology imaging workflows.
Skin Lesion Classification Using a Soft Voting Ensemble of Convolutional Neural Networks
Dec 23, 2025Skin cancer can be identified by dermoscopic examination and ocular inspection, but early detection significantly increases survival chances. Artificial intelligence (AI), using annotated skin images and Convolutional Neural Networks (CNNs), improves diagnostic accuracy. This paper presents an early skin cancer classification method using a soft voting ensemble of CNNs. In this investigation, three benchmark datasets, namely HAM10000, ISIC 2016, and ISIC 2019, were used. The process involved rebalancing, image augmentation, and filtering techniques, followed by a hybrid dual encoder for segmentation via transfer learning. Accurate segmentation focused classification models on clinically significant features, reducing background artifacts and improving accuracy. Classification was performed through an ensemble of MobileNetV2, VGG19, and InceptionV3, balancing accuracy and speed for real-world deployment. The method achieved lesion recognition accuracies of 96.32\%, 90.86\%, and 93.92\% for the three datasets. The system performance was evaluated using established skin lesion detection metrics, yielding impressive results.
LDP: Parameter-Efficient Fine-Tuning of Multimodal LLM for Medical Report Generation
Dec 11, 2025Colonoscopic polyp diagnosis is pivotal for early colorectal cancer detection, yet traditional automated reporting suffers from inconsistencies and hallucinations due to the scarcity of high-quality multimodal medical data. To bridge this gap, we propose LDP, a novel framework leveraging multimodal large language models (MLLMs) for professional polyp diagnosis report generation. Specifically, we curate MMEndo, a multimodal endoscopic dataset comprising expert-annotated colonoscopy image-text pairs. We fine-tune the Qwen2-VL-7B backbone using Parameter-Efficient Fine-Tuning (LoRA) and align it with clinical standards via Direct Preference Optimization (DPO). Extensive experiments show that our LDP outperforms existing baselines on both automated metrics and rigorous clinical expert evaluations (achieving a Physician Score of 7.2/10), significantly reducing training computational costs by 833x compared to full fine-tuning. The proposed solution offers a scalable, clinically viable path for primary healthcare, with additional validation on the IU-XRay dataset confirming its robustness.
Leveraging Machine Learning for Early Detection of Lung Diseases
Dec 27, 2025A combination of traditional image processing methods with advanced neural networks concretes a predictive and preventive healthcare paradigm. This study offers rapid, accurate, and non-invasive diagnostic solutions that can significantly impact patient outcomes, particularly in areas with limited access to radiologists and healthcare resources. In this project, deep learning methods apply in enhancing the diagnosis of respiratory diseases such as COVID-19, lung cancer, and pneumonia from chest x-rays. We trained and validated various neural network models, including CNNs, VGG16, InceptionV3, and EfficientNetB0, with high accuracy, precision, recall, and F1 scores to highlight the models' reliability and potential in real-world diagnostic applications.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Fairness Evaluation of Risk Estimation Models for Lung Cancer Screening
Dec 23, 2025Lung cancer is the leading cause of cancer-related mortality in adults worldwide. Screening high-risk individuals with annual low-dose CT (LDCT) can support earlier detection and reduce deaths, but widespread implementation may strain the already limited radiology workforce. AI models have shown potential in estimating lung cancer risk from LDCT scans. However, high-risk populations for lung cancer are diverse, and these models' performance across demographic groups remains an open question. In this study, we drew on the considerations on confounding factors and ethically significant biases outlined in the JustEFAB framework to evaluate potential performance disparities and fairness in two deep learning risk estimation models for lung cancer screening: the Sybil lung cancer risk model and the Venkadesh21 nodule risk estimator. We also examined disparities in the PanCan2b logistic regression model recommended in the British Thoracic Society nodule management guideline. Both deep learning models were trained on data from the US-based National Lung Screening Trial (NLST), and assessed on a held-out NLST validation set. We evaluated AUROC, sensitivity, and specificity across demographic subgroups, and explored potential confounding from clinical risk factors. We observed a statistically significant AUROC difference in Sybil's performance between women (0.88, 95% CI: 0.86, 0.90) and men (0.81, 95% CI: 0.78, 0.84, p < .001). At 90% specificity, Venkadesh21 showed lower sensitivity for Black (0.39, 95% CI: 0.23, 0.59) than White participants (0.69, 95% CI: 0.65, 0.73). These differences were not explained by available clinical confounders and thus may be classified as unfair biases according to JustEFAB. Our findings highlight the importance of improving and monitoring model performance across underrepresented subgroups, and further research on algorithmic fairness, in lung cancer screening.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:025
DBT-DINO: Towards Foundation model based analysis of Digital Breast Tomosynthesis
Dec 15, 2025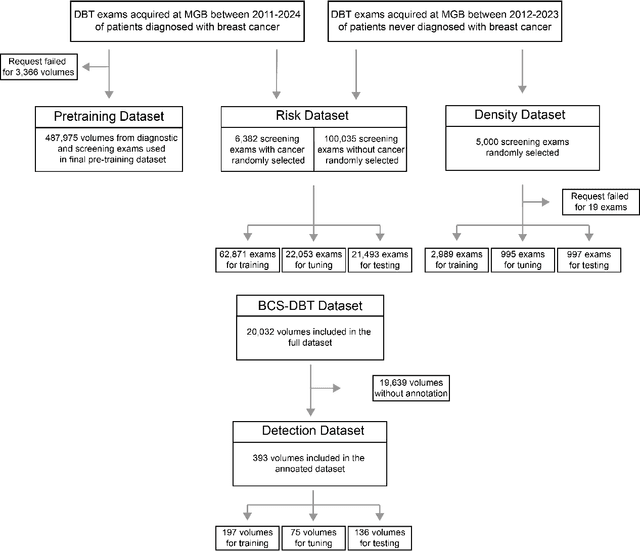
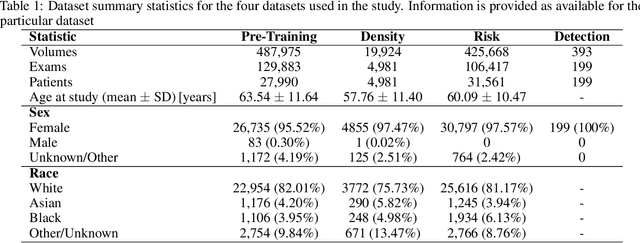
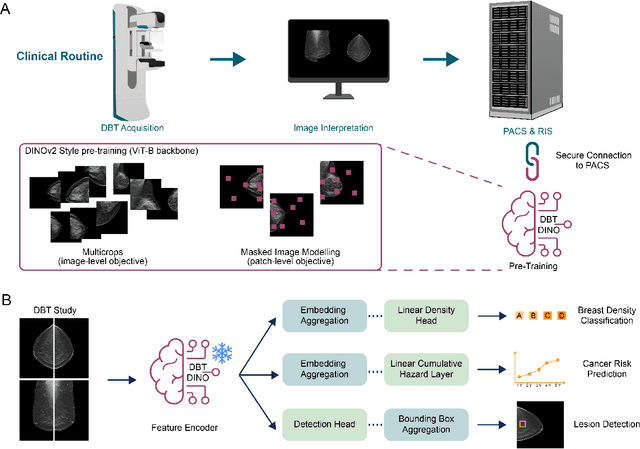
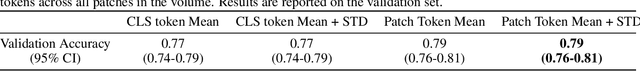
Foundation models have shown promise in medical imaging but remain underexplored for three-dimensional imaging modalities. No foundation model currently exists for Digital Breast Tomosynthesis (DBT), despite its use for breast cancer screening. To develop and evaluate a foundation model for DBT (DBT-DINO) across multiple clinical tasks and assess the impact of domain-specific pre-training. Self-supervised pre-training was performed using the DINOv2 methodology on over 25 million 2D slices from 487,975 DBT volumes from 27,990 patients. Three downstream tasks were evaluated: (1) breast density classification using 5,000 screening exams; (2) 5-year risk of developing breast cancer using 106,417 screening exams; and (3) lesion detection using 393 annotated volumes. For breast density classification, DBT-DINO achieved an accuracy of 0.79 (95\% CI: 0.76--0.81), outperforming both the MetaAI DINOv2 baseline (0.73, 95\% CI: 0.70--0.76, p<.001) and DenseNet-121 (0.74, 95\% CI: 0.71--0.76, p<.001). For 5-year breast cancer risk prediction, DBT-DINO achieved an AUROC of 0.78 (95\% CI: 0.76--0.80) compared to DINOv2's 0.76 (95\% CI: 0.74--0.78, p=.57). For lesion detection, DINOv2 achieved a higher average sensitivity of 0.67 (95\% CI: 0.60--0.74) compared to DBT-DINO with 0.62 (95\% CI: 0.53--0.71, p=.60). DBT-DINO demonstrated better performance on cancerous lesions specifically with a detection rate of 78.8\% compared to Dinov2's 77.3\%. Using a dataset of unprecedented size, we developed DBT-DINO, the first foundation model for DBT. DBT-DINO demonstrated strong performance on breast density classification and cancer risk prediction. However, domain-specific pre-training showed variable benefits on the detection task, with ImageNet baseline outperforming DBT-DINO on general lesion detection, indicating that localized detection tasks require further methodological development.
Tumor-anchored deep feature random forests for out-of-distribution detection in lung cancer segmentation
Dec 09, 2025Accurate segmentation of cancerous lesions from 3D computed tomography (CT) scans is essential for automated treatment planning and response assessment. However, even state-of-the-art models combining self-supervised learning (SSL) pretrained transformers with convolutional decoders are susceptible to out-of-distribution (OOD) inputs, generating confidently incorrect tumor segmentations, posing risks for safe clinical deployment. Existing logit-based methods suffer from task-specific model biases, while architectural enhancements to explicitly detect OOD increase parameters and computational costs. Hence, we introduce a plug-and-play and lightweight post-hoc random forests-based OOD detection framework called RF-Deep that leverages deep features with limited outlier exposure. RF-Deep enhances generalization to imaging variations by repurposing the hierarchical features from the pretrained-then-finetuned backbone encoder, providing task-relevant OOD detection by extracting the features from multiple regions of interest anchored to the predicted tumor segmentations. Hence, it scales to images of varying fields-of-view. We compared RF-Deep against existing OOD detection methods using 1,916 CT scans across near-OOD (pulmonary embolism, negative COVID-19) and far-OOD (kidney cancer, healthy pancreas) datasets. RF-Deep achieved AUROC > 93.50 for the challenging near-OOD datasets and near-perfect detection (AUROC > 99.00) for the far-OOD datasets, substantially outperforming logit-based and radiomics approaches. RF-Deep maintained similar performance consistency across networks of different depths and pretraining strategies, demonstrating its effectiveness as a lightweight, architecture-agnostic approach to enhance the reliability of tumor segmentation from CT volumes.
From SAM to DINOv2: Towards Distilling Foundation Models to Lightweight Baselines for Generalized Polyp Segmentation
Dec 10, 2025Accurate polyp segmentation during colonoscopy is critical for the early detection of colorectal cancer and still remains challenging due to significant size, shape, and color variations, and the camouflaged nature of polyps. While lightweight baseline models such as U-Net, U-Net++, and PraNet offer advantages in terms of easy deployment and low computational cost, they struggle to deal with the above issues, leading to limited segmentation performance. In contrast, large-scale vision foundation models such as SAM, DINOv2, OneFormer, and Mask2Former have exhibited impressive generalization performance across natural image domains. However, their direct transfer to medical imaging tasks (e.g., colonoscopic polyp segmentation) is not straightforward, primarily due to the scarcity of large-scale datasets and lack of domain-specific knowledge. To bridge this gap, we propose a novel distillation framework, Polyp-DiFoM, that transfers the rich representations of foundation models into lightweight segmentation baselines, allowing efficient and accurate deployment in clinical settings. In particular, we infuse semantic priors from the foundation models into canonical architectures such as U-Net and U-Net++ and further perform frequency domain encoding for enhanced distillation, corroborating their generalization capability. Extensive experiments are performed across five benchmark datasets, such as Kvasir-SEG, CVC-ClinicDB, ETIS, ColonDB, and CVC-300. Notably, Polyp-DiFoM consistently outperforms respective baseline models significantly, as well as the state-of-the-art model, with nearly 9 times reduced computation overhead. The code is available at https://github.com/lostinrepo/PolypDiFoM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge