cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
DBT-DINO: Towards Foundation model based analysis of Digital Breast Tomosynthesis
Dec 15, 2025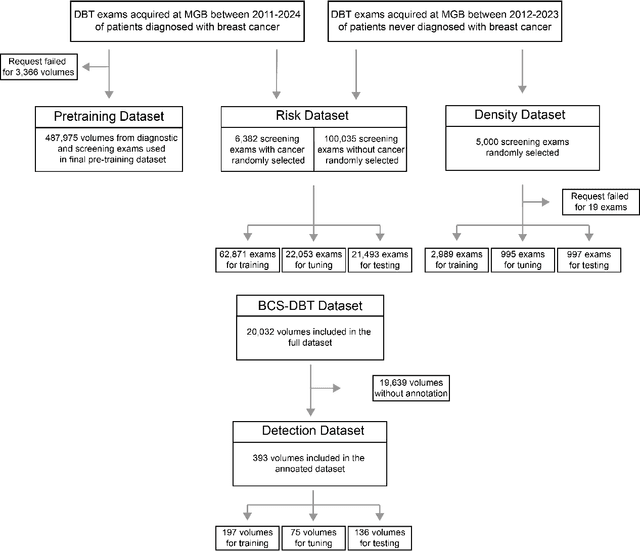
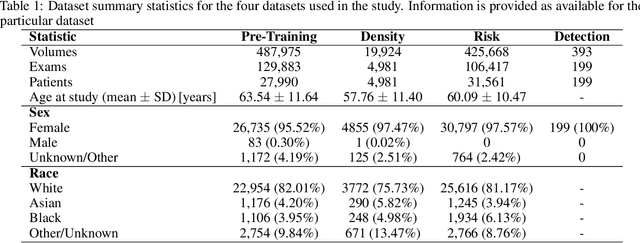
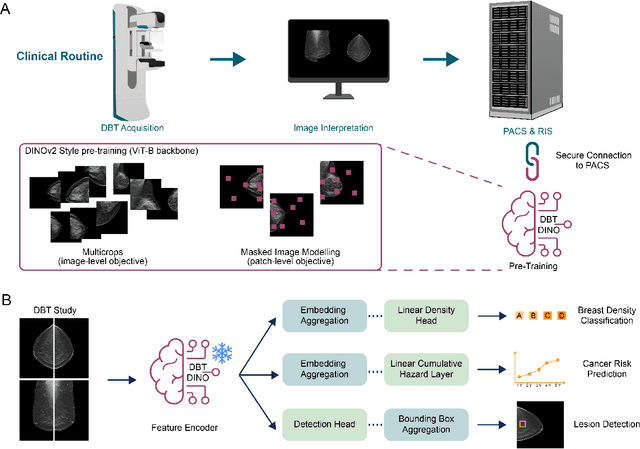
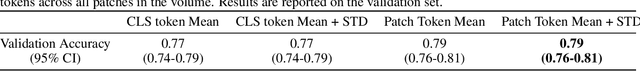
Foundation models have shown promise in medical imaging but remain underexplored for three-dimensional imaging modalities. No foundation model currently exists for Digital Breast Tomosynthesis (DBT), despite its use for breast cancer screening. To develop and evaluate a foundation model for DBT (DBT-DINO) across multiple clinical tasks and assess the impact of domain-specific pre-training. Self-supervised pre-training was performed using the DINOv2 methodology on over 25 million 2D slices from 487,975 DBT volumes from 27,990 patients. Three downstream tasks were evaluated: (1) breast density classification using 5,000 screening exams; (2) 5-year risk of developing breast cancer using 106,417 screening exams; and (3) lesion detection using 393 annotated volumes. For breast density classification, DBT-DINO achieved an accuracy of 0.79 (95\% CI: 0.76--0.81), outperforming both the MetaAI DINOv2 baseline (0.73, 95\% CI: 0.70--0.76, p<.001) and DenseNet-121 (0.74, 95\% CI: 0.71--0.76, p<.001). For 5-year breast cancer risk prediction, DBT-DINO achieved an AUROC of 0.78 (95\% CI: 0.76--0.80) compared to DINOv2's 0.76 (95\% CI: 0.74--0.78, p=.57). For lesion detection, DINOv2 achieved a higher average sensitivity of 0.67 (95\% CI: 0.60--0.74) compared to DBT-DINO with 0.62 (95\% CI: 0.53--0.71, p=.60). DBT-DINO demonstrated better performance on cancerous lesions specifically with a detection rate of 78.8\% compared to Dinov2's 77.3\%. Using a dataset of unprecedented size, we developed DBT-DINO, the first foundation model for DBT. DBT-DINO demonstrated strong performance on breast density classification and cancer risk prediction. However, domain-specific pre-training showed variable benefits on the detection task, with ImageNet baseline outperforming DBT-DINO on general lesion detection, indicating that localized detection tasks require further methodological development.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Tumor-anchored deep feature random forests for out-of-distribution detection in lung cancer segmentation
Dec 09, 2025Accurate segmentation of cancerous lesions from 3D computed tomography (CT) scans is essential for automated treatment planning and response assessment. However, even state-of-the-art models combining self-supervised learning (SSL) pretrained transformers with convolutional decoders are susceptible to out-of-distribution (OOD) inputs, generating confidently incorrect tumor segmentations, posing risks for safe clinical deployment. Existing logit-based methods suffer from task-specific model biases, while architectural enhancements to explicitly detect OOD increase parameters and computational costs. Hence, we introduce a plug-and-play and lightweight post-hoc random forests-based OOD detection framework called RF-Deep that leverages deep features with limited outlier exposure. RF-Deep enhances generalization to imaging variations by repurposing the hierarchical features from the pretrained-then-finetuned backbone encoder, providing task-relevant OOD detection by extracting the features from multiple regions of interest anchored to the predicted tumor segmentations. Hence, it scales to images of varying fields-of-view. We compared RF-Deep against existing OOD detection methods using 1,916 CT scans across near-OOD (pulmonary embolism, negative COVID-19) and far-OOD (kidney cancer, healthy pancreas) datasets. RF-Deep achieved AUROC > 93.50 for the challenging near-OOD datasets and near-perfect detection (AUROC > 99.00) for the far-OOD datasets, substantially outperforming logit-based and radiomics approaches. RF-Deep maintained similar performance consistency across networks of different depths and pretraining strategies, demonstrating its effectiveness as a lightweight, architecture-agnostic approach to enhance the reliability of tumor segmentation from CT volumes.
From SAM to DINOv2: Towards Distilling Foundation Models to Lightweight Baselines for Generalized Polyp Segmentation
Dec 10, 2025Accurate polyp segmentation during colonoscopy is critical for the early detection of colorectal cancer and still remains challenging due to significant size, shape, and color variations, and the camouflaged nature of polyps. While lightweight baseline models such as U-Net, U-Net++, and PraNet offer advantages in terms of easy deployment and low computational cost, they struggle to deal with the above issues, leading to limited segmentation performance. In contrast, large-scale vision foundation models such as SAM, DINOv2, OneFormer, and Mask2Former have exhibited impressive generalization performance across natural image domains. However, their direct transfer to medical imaging tasks (e.g., colonoscopic polyp segmentation) is not straightforward, primarily due to the scarcity of large-scale datasets and lack of domain-specific knowledge. To bridge this gap, we propose a novel distillation framework, Polyp-DiFoM, that transfers the rich representations of foundation models into lightweight segmentation baselines, allowing efficient and accurate deployment in clinical settings. In particular, we infuse semantic priors from the foundation models into canonical architectures such as U-Net and U-Net++ and further perform frequency domain encoding for enhanced distillation, corroborating their generalization capability. Extensive experiments are performed across five benchmark datasets, such as Kvasir-SEG, CVC-ClinicDB, ETIS, ColonDB, and CVC-300. Notably, Polyp-DiFoM consistently outperforms respective baseline models significantly, as well as the state-of-the-art model, with nearly 9 times reduced computation overhead. The code is available at https://github.com/lostinrepo/PolypDiFoM.
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
General OOD Detection via Model-aware and Subspace-aware Variable Priority
Dec 15, 2025Out-of-distribution (OOD) detection is essential for determining when a supervised model encounters inputs that differ meaningfully from its training distribution. While widely studied in classification, OOD detection for regression and survival analysis remains limited due to the absence of discrete labels and the challenge of quantifying predictive uncertainty. We introduce a framework for OOD detection that is simultaneously model aware and subspace aware, and that embeds variable prioritization directly into the detection step. The method uses the fitted predictor to construct localized neighborhoods around each test case that emphasize the features driving the model's learned relationship and downweight directions that are less relevant to prediction. It produces OOD scores without relying on global distance metrics or estimating the full feature density. The framework is applicable across outcome types, and in our implementation we use random forests, where the rule structure yields transparent neighborhoods and effective scoring. Experiments on synthetic and real data benchmarks designed to isolate functional shifts show consistent improvements over existing methods. We further demonstrate the approach in an esophageal cancer survival study, where distribution shifts related to lymphadenectomy identify patterns relevant to surgical guidelines.
Configurable γ Photon Spectrometer to Enable Precision Radioguided Tumor Resection
Dec 16, 2025Surgical tumor resection aims to remove all cancer cells in the tumor margin and at centimeter-scale depths below the tissue surface. During surgery, microscopic clusters of disease are intraoperatively difficult to visualize and are often left behind, significantly increasing the risk of cancer recurrence. Radioguided surgery (RGS) has shown the ability to selectively tag cancer cells with gamma (γ) photon emitting radioisotopes to identify them, but require a mm-scale γ photon spectrometer to localize the position of these cells in the tissue margin (i.e., a function of incident γ photon energy) with high specificity. Here we present a 9.9 mm2 integrated circuit (IC)-based γ spectrometer implemented in 180 nm CMOS, to enable the measurement of single γ photons and their incident energy with sub-keV energy resolution. We use small 2 2 um reverse-biased diodes that have low depletion region capacitance, and therefore produce millivolt-scale voltage signals in response to the small charge generated by incident γ photons. A low-power energy spectrometry method is implemented by measuring the decay time it takes for the generated voltage signal to settle back to DC after a γ detection event, instead of measuring the voltage drop directly. This spectrometry method is implemented in three different pixel architectures that allow for configurable pixel sensitivity, energy-resolution, and energy dynamic range based on the widely heterogenous surgical and patient presentation in RGS. The spectrometer was tested with three common γ-emitting radioisotopes (64Cu, 133Ba, 177Lu), and is able to resolve activities down to 1 uCi with sub-keV energy resolution and 1.315 MeV energy dynamic range, using 5-minute acquisitions.
Topological Conditioning for Mammography Models via a Stable Wavelet-Persistence Vectorization
Dec 10, 2025Breast cancer is the most commonly diagnosed cancer in women and a leading cause of cancer death worldwide. Screening mammography reduces mortality, yet interpretation still suffers from substantial false negatives and false positives, and model accuracy often degrades when deployed across scanners, modalities, and patient populations. We propose a simple conditioning signal aimed at improving external performance based on a wavelet based vectorization of persistent homology. Using topological data analysis, we summarize image structure that persists across intensity thresholds and convert this information into spatial, multi scale maps that are provably stable to small intensity perturbations. These maps are integrated into a two stage detection pipeline through input level channel concatenation. The model is trained and validated on the CBIS DDSM digitized film mammography cohort from the United States and evaluated on two independent full field digital mammography cohorts from Portugal (INbreast) and China (CMMD), with performance reported at the patient level. On INbreast, augmenting ConvNeXt Tiny with wavelet persistence channels increases patient level AUC from 0.55 to 0.75 under a limited training budget.
Explaining Digital Pathology Models via Clustering Activations
Nov 18, 2025We present a clustering-based explainability technique for digital pathology models based on convolutional neural networks. Unlike commonly used methods based on saliency maps, such as occlusion, GradCAM, or relevance propagation, which highlight regions that contribute the most to the prediction for a single slide, our method shows the global behaviour of the model under consideration, while also providing more fine-grained information. The result clusters can be visualised not only to understand the model, but also to increase confidence in its operation, leading to faster adoption in clinical practice. We also evaluate the performance of our technique on an existing model for detecting prostate cancer, demonstrating its usefulness.
From 2D to 3D Without Extra Baggage: Data-Efficient Cancer Detection in Digital Breast Tomosynthesis
Nov 13, 2025



Digital Breast Tomosynthesis (DBT) enhances finding visibility for breast cancer detection by providing volumetric information that reduces the impact of overlapping tissues; however, limited annotated data has constrained the development of deep learning models for DBT. To address data scarcity, existing methods attempt to reuse 2D full-field digital mammography (FFDM) models by either flattening DBT volumes or processing slices individually, thus discarding volumetric information. Alternatively, 3D reasoning approaches introduce complex architectures that require more DBT training data. Tackling these drawbacks, we propose M&M-3D, an architecture that enables learnable 3D reasoning while remaining parameter-free relative to its FFDM counterpart, M&M. M&M-3D constructs malignancy-guided 3D features, and 3D reasoning is learned through repeatedly mixing these 3D features with slice-level information. This is achieved by modifying operations in M&M without adding parameters, thus enabling direct weight transfer from FFDM. Extensive experiments show that M&M-3D surpasses 2D projection and 3D slice-based methods by 11-54% for localization and 3-10% for classification. Additionally, M&M-3D outperforms complex 3D reasoning variants by 20-47% for localization and 2-10% for classification in the low-data regime, while matching their performance in high-data regime. On the popular BCS-DBT benchmark, M&M-3D outperforms previous top baseline by 4% for classification and 10% for localization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge