Image To Image Translation
Image-to-image translation is the process of converting an image from one domain to another using deep learning techniques.
Papers and Code
Parameter Efficient Multimodal Instruction Tuning for Romanian Vision Language Models
Dec 16, 2025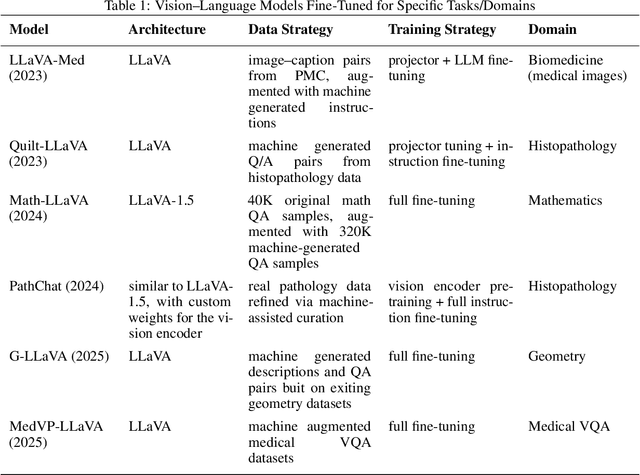
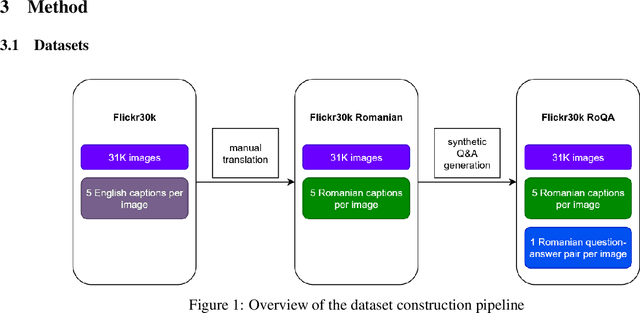
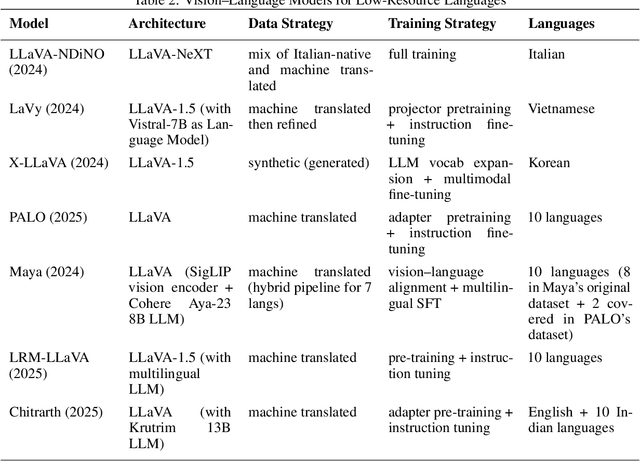
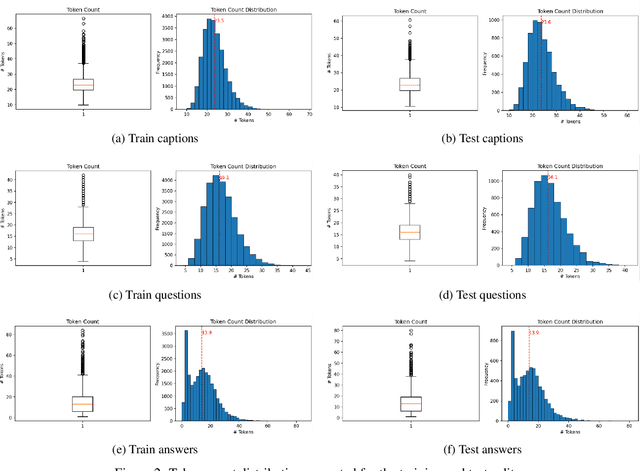
Focusing on low-resource languages is an essential step toward democratizing generative AI. In this work, we contribute to reducing the multimodal NLP resource gap for Romanian. We translate the widely known Flickr30k dataset into Romanian and further extend it for visual question answering by leveraging open-source LLMs. We demonstrate the usefulness of our datasets by fine-tuning open-source VLMs on Romanian visual question answering. We select VLMs from three widely used model families: LLaMA 3.2, LLaVA 1.6, and Qwen2. For fine-tuning, we employ the parameter-efficient LoRA method. Our models show improved Romanian capabilities in visual QA, as well as on tasks they were not trained on, such as Romanian image description generation. The seven-billion-parameter Qwen2-VL-RoVQA obtains top scores on both tasks, with improvements of +6.05% and +2.61% in BERTScore F1 over its original version. Finally, the models show substantial reductions in grammatical errors compared to their original forms, indicating improvements not only in language understanding but also in Romanian fluency.
AMD-HookNet++: Evolution of AMD-HookNet with Hybrid CNN-Transformer Feature Enhancement for Glacier Calving Front Segmentation
Dec 16, 2025The dynamics of glaciers and ice shelf fronts significantly impact the mass balance of ice sheets and coastal sea levels. To effectively monitor glacier conditions, it is crucial to consistently estimate positional shifts of glacier calving fronts. AMD-HookNet firstly introduces a pure two-branch convolutional neural network (CNN) for glacier segmentation. Yet, the local nature and translational invariance of convolution operations, while beneficial for capturing low-level details, restricts the model ability to maintain long-range dependencies. In this study, we propose AMD-HookNet++, a novel advanced hybrid CNN-Transformer feature enhancement method for segmenting glaciers and delineating calving fronts in synthetic aperture radar images. Our hybrid structure consists of two branches: a Transformer-based context branch to capture long-range dependencies, which provides global contextual information in a larger view, and a CNN-based target branch to preserve local details. To strengthen the representation of the connected hybrid features, we devise an enhanced spatial-channel attention module to foster interactions between the hybrid CNN-Transformer branches through dynamically adjusting the token relationships from both spatial and channel perspectives. Additionally, we develop a pixel-to-pixel contrastive deep supervision to optimize our hybrid model by integrating pixelwise metric learning into glacier segmentation. Through extensive experiments and comprehensive quantitative and qualitative analyses on the challenging glacier segmentation benchmark dataset CaFFe, we show that AMD-HookNet++ sets a new state of the art with an IoU of 78.2 and a HD95 of 1,318 m, while maintaining a competitive MDE of 367 m. More importantly, our hybrid model produces smoother delineations of calving fronts, resolving the issue of jagged edges typically seen in pure Transformer-based approaches.
Towards Scalable Pre-training of Visual Tokenizers for Generation
Dec 15, 2025The quality of the latent space in visual tokenizers (e.g., VAEs) is crucial for modern generative models. However, the standard reconstruction-based training paradigm produces a latent space that is biased towards low-level information, leading to a foundation flaw: better pixel-level accuracy does not lead to higher-quality generation. This implies that pouring extensive compute into visual tokenizer pre-training translates poorly to improved performance in generation. We identify this as the ``pre-training scaling problem`` and suggest a necessary shift: to be effective for generation, a latent space must concisely represent high-level semantics. We present VTP, a unified visual tokenizer pre-training framework, pioneering the joint optimization of image-text contrastive, self-supervised, and reconstruction losses. Our large-scale study reveals two principal findings: (1) understanding is a key driver of generation, and (2) much better scaling properties, where generative performance scales effectively with compute, parameters, and data allocated to the pretraining of the visual tokenizer. After large-scale pre-training, our tokenizer delivers a competitive profile (78.2 zero-shot accuracy and 0.36 rFID on ImageNet) and 4.1 times faster convergence on generation compared to advanced distillation methods. More importantly, it scales effectively: without modifying standard DiT training specs, solely investing more FLOPS in pretraining VTP achieves 65.8\% FID improvement in downstream generation, while conventional autoencoder stagnates very early at 1/10 FLOPS. Our pre-trained models are available at https://github.com/MiniMax-AI/VTP.
Cauchy-Schwarz Fairness Regularizer
Dec 10, 2025



Group fairness in machine learning is often enforced by adding a regularizer that reduces the dependence between model predictions and sensitive attributes. However, existing regularizers are built on heterogeneous distance measures and design choices, which makes their behavior hard to reason about and their performance inconsistent across tasks. This raises a basic question: what properties make a good fairness regularizer? We address this question by first organizing existing in-process methods into three families: (i) matching prediction statistics across sensitive groups, (ii) aligning latent representations, and (iii) directly minimizing dependence between predictions and sensitive attributes. Through this lens, we identify desirable properties of the underlying distance measure, including tight generalization bounds, robustness to scale differences, and the ability to handle arbitrary prediction distributions. Motivated by these properties, we propose a Cauchy-Schwarz (CS) fairness regularizer that penalizes the empirical CS divergence between prediction distributions conditioned on sensitive groups. Under a Gaussian comparison, we show that CS divergence yields a tighter bound than Kullback-Leibler divergence, Maximum Mean Discrepancy, and the mean disparity used in Demographic Parity, and we discuss how these advantages translate to a distribution-free, kernel-based estimator that naturally extends to multiple sensitive attributes. Extensive experiments on four tabular benchmarks and one image dataset demonstrate that the proposed CS regularizer consistently improves Demographic Parity and Equal Opportunity metrics while maintaining competitive accuracy, and achieves a more stable utility-fairness trade-off across hyperparameter settings compared to prior regularizers.
Multi-scale Attention-Guided Intrinsic Decomposition and Rendering Pass Prediction for Facial Images
Dec 18, 2025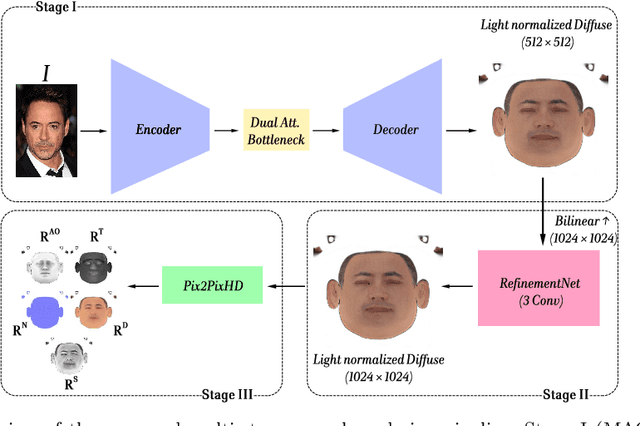
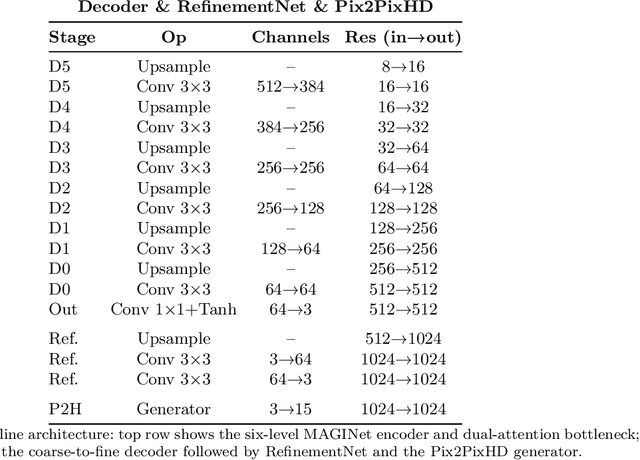
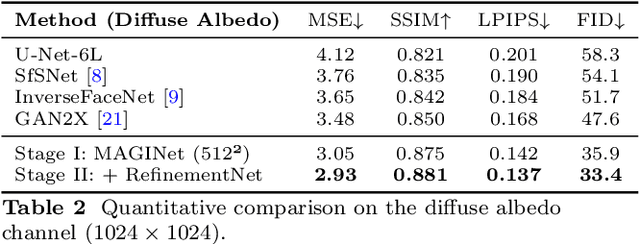
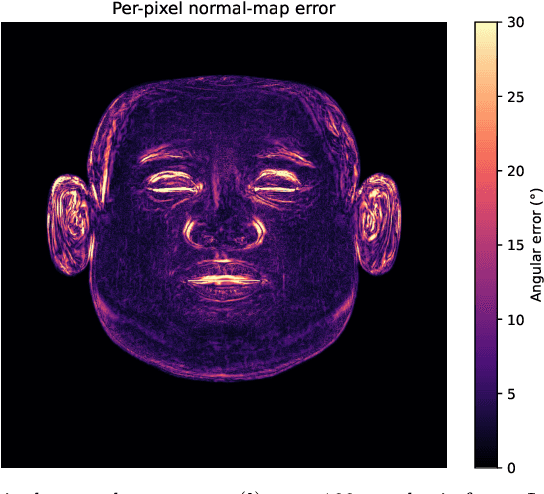
Accurate intrinsic decomposition of face images under unconstrained lighting is a prerequisite for photorealistic relighting, high-fidelity digital doubles, and augmented-reality effects. This paper introduces MAGINet, a Multi-scale Attention-Guided Intrinsics Network that predicts a $512\times512$ light-normalized diffuse albedo map from a single RGB portrait. MAGINet employs hierarchical residual encoding, spatial-and-channel attention in a bottleneck, and adaptive multi-scale feature fusion in the decoder, yielding sharper albedo boundaries and stronger lighting invariance than prior U-Net variants. The initial albedo prediction is upsampled to $1024\times1024$ and refined by a lightweight three-layer CNN (RefinementNet). Conditioned on this refined albedo, a Pix2PixHD-based translator then predicts a comprehensive set of five additional physically based rendering passes: ambient occlusion, surface normal, specular reflectance, translucency, and raw diffuse colour (with residual lighting). Together with the refined albedo, these six passes form the complete intrinsic decomposition. Trained with a combination of masked-MSE, VGG, edge, and patch-LPIPS losses on the FFHQ-UV-Intrinsics dataset, the full pipeline achieves state-of-the-art performance for diffuse albedo estimation and demonstrates significantly improved fidelity for the complete rendering stack compared to prior methods. The resulting passes enable high-quality relighting and material editing of real faces.
Bitbox: Behavioral Imaging Toolbox for Computational Analysis of Behavior from Videos
Dec 19, 2025



Computational measurement of human behavior from video has recently become feasible due to major advances in AI. These advances now enable granular and precise quantification of facial expression, head movement, body action, and other behavioral modalities and are increasingly used in psychology, psychiatry, neuroscience, and mental health research. However, mainstream adoption remains slow. Most existing methods and software are developed for engineering audiences, require specialized software stacks, and fail to provide behavioral measurements at a level directly useful for hypothesis-driven research. As a result, there is a large barrier to entry for researchers who wish to use modern, AI-based tools in their work. We introduce Bitbox, an open-source toolkit designed to remove this barrier and make advanced computational analysis directly usable by behavioral scientists and clinical researchers. Bitbox is guided by principles of reproducibility, modularity, and interpretability. It provides a standardized interface for extracting high-level behavioral measurements from video, leveraging multiple face, head, and body processors. The core modules have been tested and validated on clinical samples and are designed so that new measures can be added with minimal effort. Bitbox is intended to serve both sides of the translational gap. It gives behavioral researchers access to robust, high-level behavioral metrics without requiring engineering expertise, and it provides computer scientists a practical mechanism for disseminating methods to domains where their impact is most needed. We expect that Bitbox will accelerate integration of computational behavioral measurement into behavioral, clinical, and mental health research. Bitbox has been designed from the beginning as a community-driven effort that will evolve through contributions from both method developers and domain scientists.
Uncertainty-Guided Selective Adaptation Enables Cross-Platform Predictive Fluorescence Microscopy
Nov 15, 2025Deep learning is transforming microscopy, yet models often fail when applied to images from new instruments or acquisition settings. Conventional adversarial domain adaptation (ADDA) retrains entire networks, often disrupting learned semantic representations. Here, we overturn this paradigm by showing that adapting only the earliest convolutional layers, while freezing deeper layers, yields reliable transfer. Building on this principle, we introduce Subnetwork Image Translation ADDA with automatic depth selection (SIT-ADDA-Auto), a self-configuring framework that integrates shallow-layer adversarial alignment with predictive uncertainty to automatically select adaptation depth without target labels. We demonstrate robustness via multi-metric evaluation, blinded expert assessment, and uncertainty-depth ablations. Across exposure and illumination shifts, cross-instrument transfer, and multiple stains, SIT-ADDA improves reconstruction and downstream segmentation over full-encoder adaptation and non-adversarial baselines, with reduced drift of semantic features. Our results provide a design rule for label-free adaptation in microscopy and a recipe for field settings; the code is publicly available.
DINOv3-Guided Cross Fusion Framework for Semantic-aware CT generation from MRI and CBCT
Nov 15, 2025



Generating synthetic CT images from CBCT or MRI has a potential for efficient radiation dose planning and adaptive radiotherapy. However, existing CNN-based models lack global semantic understanding, while Transformers often overfit small medical datasets due to high model capacity and weak inductive bias. To address these limitations, we propose a DINOv3-Guided Cross Fusion (DGCF) framework that integrates a frozen self-supervised DINOv3 Transformer with a trainable CNN encoder-decoder. It hierarchically fuses global representation of Transformer and local features of CNN via a learnable cross fusion module, achieving balanced local appearance and contextual representation. Furthermore, we introduce a Multi-Level DINOv3 Perceptual (MLDP) loss that encourages semantic similarity between synthetic CT and the ground truth in DINOv3's feature space. Experiments on the SynthRAD2023 pelvic dataset demonstrate that DGCF achieved state-of-the-art performance in terms of MS-SSIM, PSNR and segmentation-based metrics on both MRI$\rightarrow$CT and CBCT$\rightarrow$CT translation tasks. To the best of our knowledge, this is the first work to employ DINOv3 representations for medical image translation, highlighting the potential of self-supervised Transformer guidance for semantic-aware CT synthesis. The code is available at https://github.com/HiLab-git/DGCF.
Electromagnetic Quantitative Inversion for Translationally Moving Targets via Phase Correlation Registration of Back-Projection Images
Nov 19, 2025A novel electromagnetic quantitative inversion scheme for translationally moving targets via phase correlation registration of back-projection (BP) images is proposed. Based on a time division multiplexing multiple-input multiple-output (TDM-MIMO) radar architecture, the scheme first achieves high-precision relative positioning of the target, then applies relative motion compensation to perform iterative inversion on multi-cycle MIMO measurement data, thereby reconstructing the target's electromagnetic parameters. As a general framework compatible with other mainstream inversion algorithms, we exemplify our approach by incorporating the classical cross-correlated contrast source inversion (CC-CSI) into iterative optimization step of the scheme, resulting in a new algorithm termed RMC-CC-CSI. Numerical and experimental results demonstrate that RMC-CC-CSI offers accelerated convergence, enhanced reconstruction fidelity, and improved noise immunity over conventional CC-CSI for stationary targets despite increased computational cost.
NAP3D: NeRF Assisted 3D-3D Pose Alignment for Autonomous Vehicles
Dec 17, 2025Accurate localization is essential for autonomous vehicles, yet sensor noise and drift over time can lead to significant pose estimation errors, particularly in long-horizon environments. A common strategy for correcting accumulated error is visual loop closure in SLAM, which adjusts the pose graph when the agent revisits previously mapped locations. These techniques typically rely on identifying visual mappings between the current view and previously observed scenes and often require fusing data from multiple sensors. In contrast, this work introduces NeRF-Assisted 3D-3D Pose Alignment (NAP3D), a complementary approach that leverages 3D-3D correspondences between the agent's current depth image and a pre-trained Neural Radiance Field (NeRF). By directly aligning 3D points from the observed scene with synthesized points from the NeRF, NAP3D refines the estimated pose even from novel viewpoints, without relying on revisiting previously observed locations. This robust 3D-3D formulation provides advantages over conventional 2D-3D localization methods while remaining comparable in accuracy and applicability. Experiments demonstrate that NAP3D achieves camera pose correction within 5 cm on a custom dataset, robustly outperforming a 2D-3D Perspective-N-Point baseline. On TUM RGB-D, NAP3D consistently improves 3D alignment RMSE by approximately 6 cm compared to this baseline given varying noise, despite PnP achieving lower raw rotation and translation parameter error in some regimes, highlighting NAP3D's improved geometric consistency in 3D space. By providing a lightweight, dataset-agnostic tool, NAP3D complements existing SLAM and localization pipelines when traditional loop closure is unavailable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge