Zhehua Mao
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
Automated Surgical Skill Assessment in Endoscopic Pituitary Surgery using Real-time Instrument Tracking on a High-fidelity Bench-top Phantom
Sep 25, 2024Abstract:Improved surgical skill is generally associated with improved patient outcomes, although assessment is subjective; labour-intensive; and requires domain specific expertise. Automated data driven metrics can alleviate these difficulties, as demonstrated by existing machine learning instrument tracking models in minimally invasive surgery. However, these models have been tested on limited datasets of laparoscopic surgery, with a focus on isolated tasks and robotic surgery. In this paper, a new public dataset is introduced, focusing on simulated surgery, using the nasal phase of endoscopic pituitary surgery as an exemplar. Simulated surgery allows for a realistic yet repeatable environment, meaning the insights gained from automated assessment can be used by novice surgeons to hone their skills on the simulator before moving to real surgery. PRINTNet (Pituitary Real-time INstrument Tracking Network) has been created as a baseline model for this automated assessment. Consisting of DeepLabV3 for classification and segmentation; StrongSORT for tracking; and the NVIDIA Holoscan SDK for real-time performance, PRINTNet achieved 71.9% Multiple Object Tracking Precision running at 22 Frames Per Second. Using this tracking output, a Multilayer Perceptron achieved 87% accuracy in predicting surgical skill level (novice or expert), with the "ratio of total procedure time to instrument visible time" correlated with higher surgical skill. This therefore demonstrates the feasibility of automated surgical skill assessment in simulated endoscopic pituitary surgery. The new publicly available dataset can be found here: https://doi.org/10.5522/04/26511049.
Direct Simultaneous Multi-Image Registration
May 21, 2021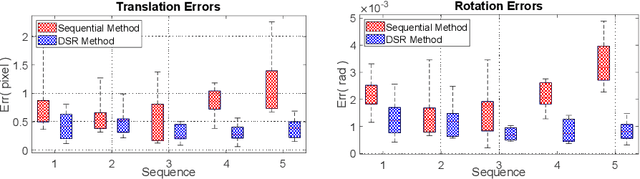
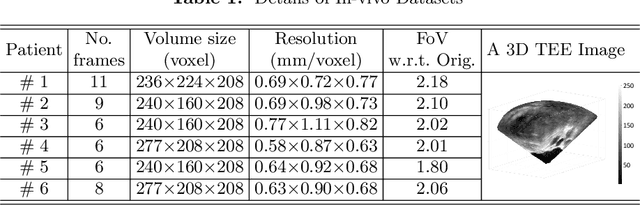
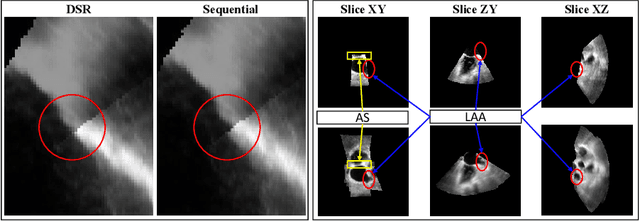
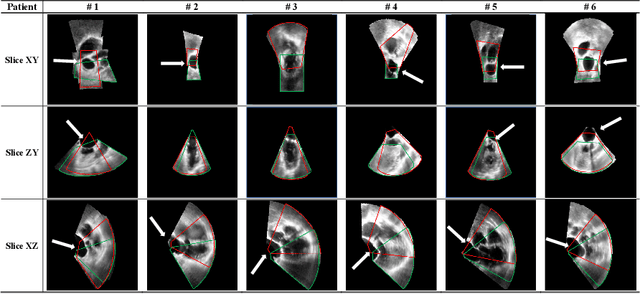
Abstract:This paper presents a novel algorithm that registers a collection of mono-modal 3D images in a simultaneous fashion, named as Direct Simultaneous Registration (DSR). The algorithm optimizes global poses of local frames directly based on the intensities of images (without extracting features from the images). To obtain the optimal result, we start with formulating a Direct Bundle Adjustment (DBA) problem which jointly optimizes pose parameters of local frames and intensities of panoramic image. By proving the independence of the pose from panoramic image in the iterative process, DSR is proposed and proved to be able to generate the same optimal poses as DBA, but without optimizing the intensities of the panoramic image. The proposed DSR method is particularly suitable in mono-modal registration and in the scenarios where distinct features are not available, such as Transesophageal Echocardiography (TEE) images. The proposed method is validated via simulated and in-vivo 3D TEE images. It is shown that the proposed method outperforms conventional sequential registration method in terms of accuracy and the obtained results can produce good alignment in in-vivo images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge