Raffaele Napolitano
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
Measuring proximity to standard planes during fetal brain ultrasound scanning
Apr 10, 2024



Abstract:This paper introduces a novel pipeline designed to bring ultrasound (US) plane pose estimation closer to clinical use for more effective navigation to the standard planes (SPs) in the fetal brain. We propose a semi-supervised segmentation model utilizing both labeled SPs and unlabeled 3D US volume slices. Our model enables reliable segmentation across a diverse set of fetal brain images. Furthermore, the model incorporates a classification mechanism to identify the fetal brain precisely. Our model not only filters out frames lacking the brain but also generates masks for those containing it, enhancing the relevance of plane pose regression in clinical settings. We focus on fetal brain navigation from 2D ultrasound (US) video analysis and combine this model with a US plane pose regression network to provide sensorless proximity detection to SPs and non-SPs planes; we emphasize the importance of proximity detection to SPs for guiding sonographers, offering a substantial advantage over traditional methods by allowing earlier and more precise adjustments during scanning. We demonstrate the practical applicability of our approach through validation on real fetal scan videos obtained from sonographers of varying expertise levels. Our findings demonstrate the potential of our approach to complement existing fetal US technologies and advance prenatal diagnostic practices.
AutoFB: Automating Fetal Biometry Estimation from Standard Ultrasound Planes
Jul 12, 2021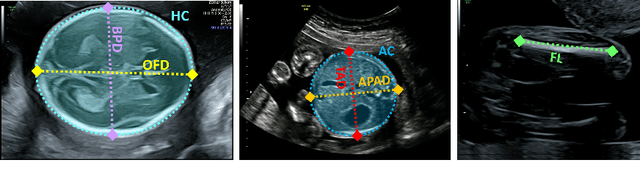
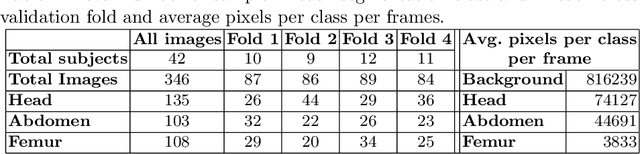
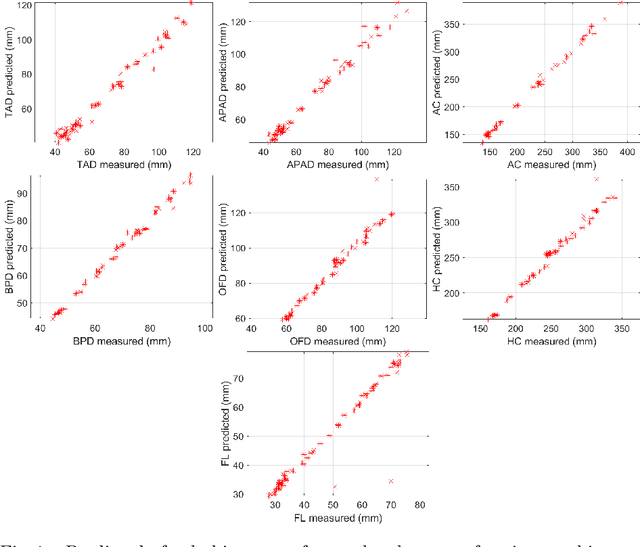
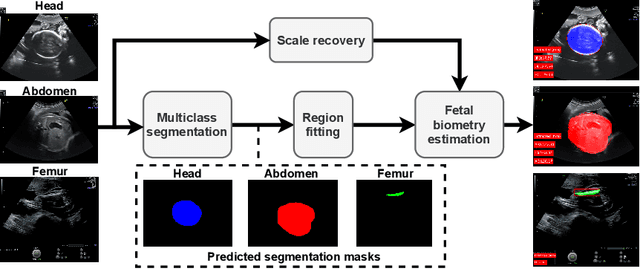
Abstract:During pregnancy, ultrasound examination in the second trimester can assess fetal size according to standardized charts. To achieve a reproducible and accurate measurement, a sonographer needs to identify three standard 2D planes of the fetal anatomy (head, abdomen, femur) and manually mark the key anatomical landmarks on the image for accurate biometry and fetal weight estimation. This can be a time-consuming operator-dependent task, especially for a trainee sonographer. Computer-assisted techniques can help in automating the fetal biometry computation process. In this paper, we present a unified automated framework for estimating all measurements needed for the fetal weight assessment. The proposed framework semantically segments the key fetal anatomies using state-of-the-art segmentation models, followed by region fitting and scale recovery for the biometry estimation. We present an ablation study of segmentation algorithms to show their robustness through 4-fold cross-validation on a dataset of 349 ultrasound standard plane images from 42 pregnancies. Moreover, we show that the network with the best segmentation performance tends to be more accurate for biometry estimation. Furthermore, we demonstrate that the error between clinically measured and predicted fetal biometry is lower than the permissible error during routine clinical measurements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge