Xing Lu
Automating Modelica Module Generation Using Large Language Models: A Case Study on Building Control Description Language
Sep 18, 2025Abstract:Dynamic energy systems and controls require advanced modeling frameworks to design and test supervisory and fault tolerant strategies. Modelica is a widely used equation based language, but developing control modules is labor intensive and requires specialized expertise. This paper examines the use of large language models (LLMs) to automate the generation of Control Description Language modules in the Building Modelica Library as a case study. We developed a structured workflow that combines standardized prompt scaffolds, library aware grounding, automated compilation with OpenModelica, and human in the loop evaluation. Experiments were carried out on four basic logic tasks (And, Or, Not, and Switch) and five control modules (chiller enable/disable, bypass valve control, cooling tower fan speed, plant requests, and relief damper control). The results showed that GPT 4o failed to produce executable Modelica code in zero shot mode, while Claude Sonnet 4 achieved up to full success for basic logic blocks with carefully engineered prompts. For control modules, success rates reached 83 percent, and failed outputs required medium level human repair (estimated one to eight hours). Retrieval augmented generation often produced mismatches in module selection (for example, And retrieved as Or), while a deterministic hard rule search strategy avoided these errors. Human evaluation also outperformed AI evaluation, since current LLMs cannot assess simulation results or validate behavioral correctness. Despite these limitations, the LLM assisted workflow reduced the average development time from 10 to 20 hours down to 4 to 6 hours per module, corresponding to 40 to 60 percent time savings. These results highlight both the potential and current limitations of LLM assisted Modelica generation, and point to future research in pre simulation validation, stronger grounding, and closed loop evaluation.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023



Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
Weakly Supervised Contrastive Learning for Chest X-Ray Report Generation
Sep 25, 2021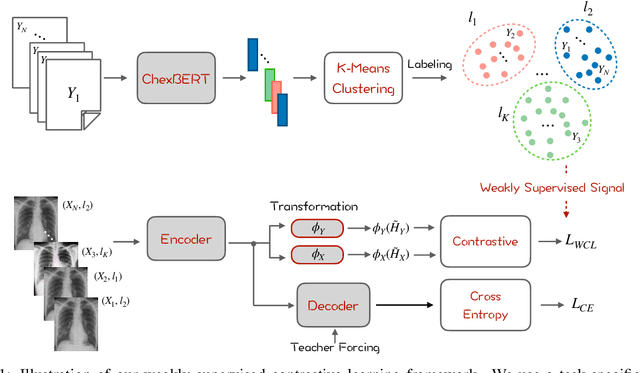
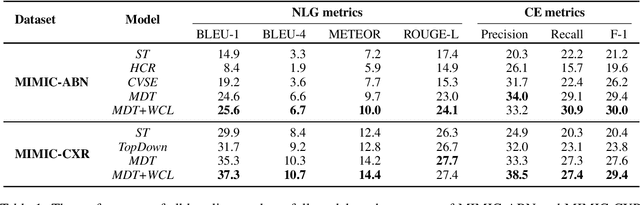
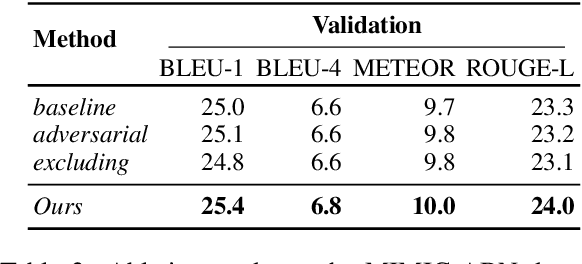
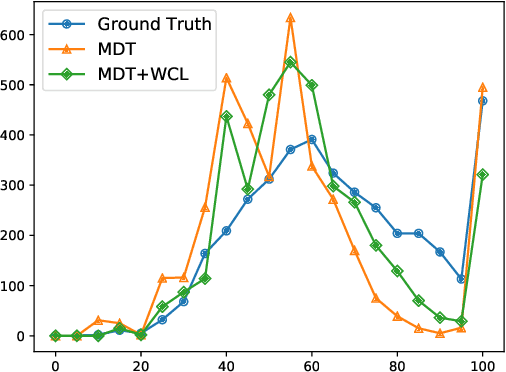
Abstract:Radiology report generation aims at generating descriptive text from radiology images automatically, which may present an opportunity to improve radiology reporting and interpretation. A typical setting consists of training encoder-decoder models on image-report pairs with a cross entropy loss, which struggles to generate informative sentences for clinical diagnoses since normal findings dominate the datasets. To tackle this challenge and encourage more clinically-accurate text outputs, we propose a novel weakly supervised contrastive loss for medical report generation. Experimental results demonstrate that our method benefits from contrasting target reports with incorrect but semantically-close ones. It outperforms previous work on both clinical correctness and text generation metrics for two public benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge