Toan Duc Bui
Senior Member, IEEE
Offset Curves Loss for Imbalanced Problem in Medical Segmentation
Dec 04, 2020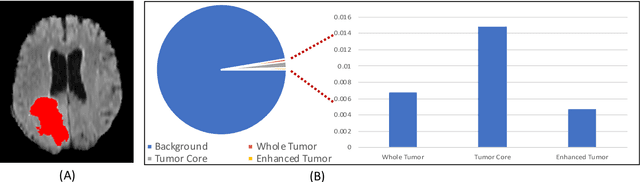
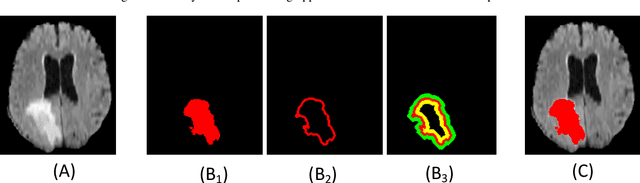
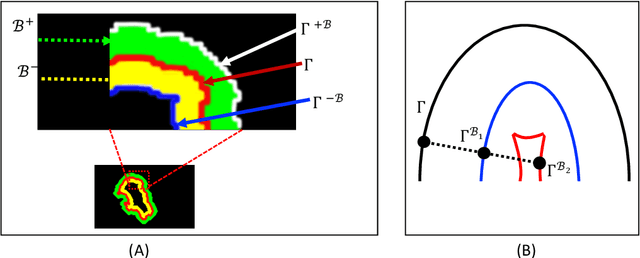
Abstract:Medical image segmentation has played an important role in medical analysis and widely developed for many clinical applications. Deep learning-based approaches have achieved high performance in semantic segmentation but they are limited to pixel-wise setting and imbalanced classes data problem. In this paper, we tackle those limitations by developing a new deep learning-based model which takes into account both higher feature level i.e. region inside contour, intermediate feature level i.e. offset curves around the contour and lower feature level i.e. contour. Our proposed Offset Curves (OsC) loss consists of three main fitting terms. The first fitting term focuses on pixel-wise level segmentation whereas the second fitting term acts as attention model which pays attention to the area around the boundaries (offset curves). The third terms plays a role as regularization term which takes the length of boundaries into account. We evaluate our proposed OsC loss on both 2D network and 3D network. Two common medical datasets, i.e. retina DRIVE and brain tumor BRATS 2018 datasets are used to benchmark our proposed loss performance. The experiments have shown that our proposed OsC loss function outperforms other mainstream loss functions such as Cross-Entropy, Dice, Focal on the most common segmentation networks Unet, FCN.
Flow-based Deformation Guidance for Unpaired Multi-Contrast MRI Image-to-Image Translation
Dec 03, 2020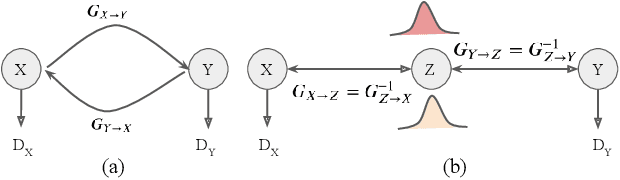
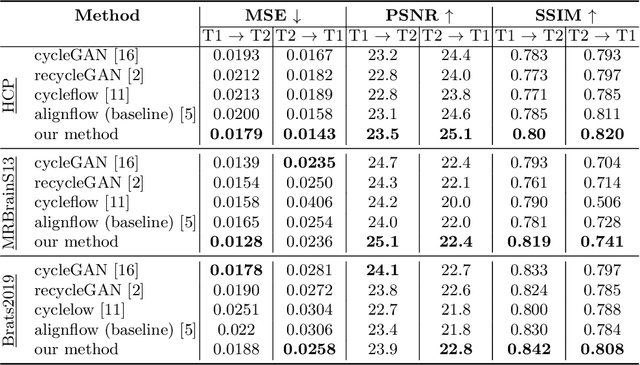
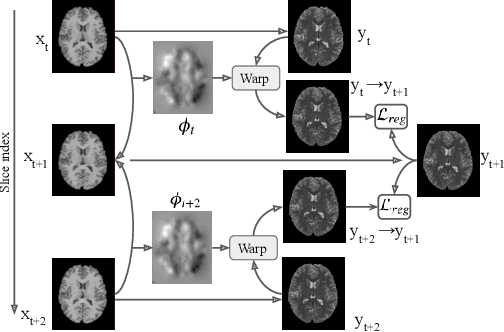
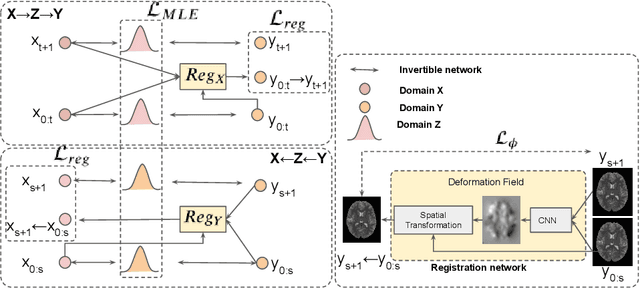
Abstract:Image synthesis from corrupted contrasts increases the diversity of diagnostic information available for many neurological diseases. Recently the image-to-image translation has experienced significant levels of interest within medical research, beginning with the successful use of the Generative Adversarial Network (GAN) to the introduction of cyclic constraint extended to multiple domains. However, in current approaches, there is no guarantee that the mapping between the two image domains would be unique or one-to-one. In this paper, we introduce a novel approach to unpaired image-to-image translation based on the invertible architecture. The invertible property of the flow-based architecture assures a cycle-consistency of image-to-image translation without additional loss functions. We utilize the temporal information between consecutive slices to provide more constraints to the optimization for transforming one domain to another in unpaired volumetric medical images. To capture temporal structures in the medical images, we explore the displacement between the consecutive slices using a deformation field. In our approach, the deformation field is used as a guidance to keep the translated slides realistic and consistent across the translation. The experimental results have shown that the synthesized images using our proposed approach are able to archive a competitive performance in terms of mean squared error, peak signal-to-noise ratio, and structural similarity index when compared with the existing deep learning-based methods on three standard datasets, i.e. HCP, MRBrainS13, and Brats2019.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020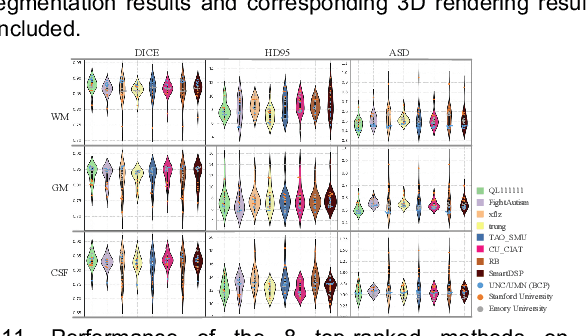
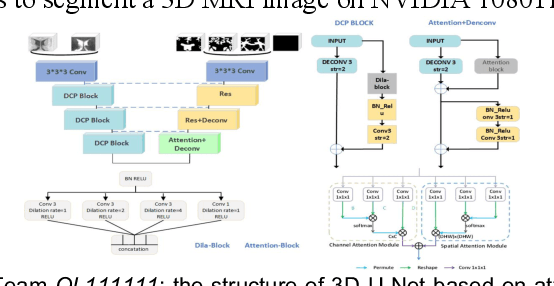
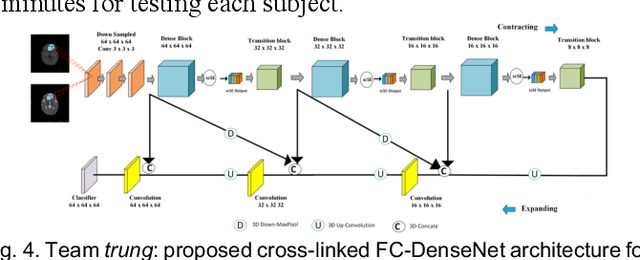
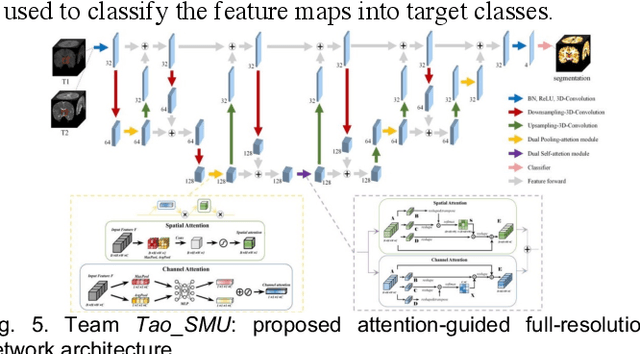
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
3D Densely Convolutional Networks for Volumetric Segmentation
Sep 13, 2017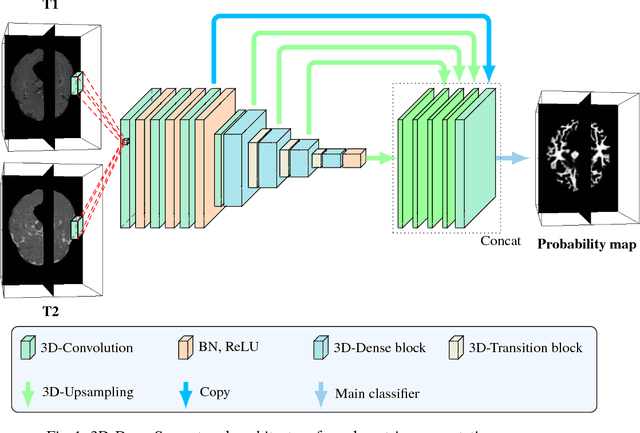
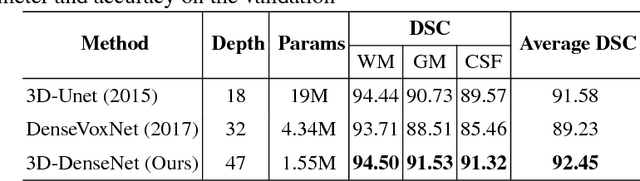
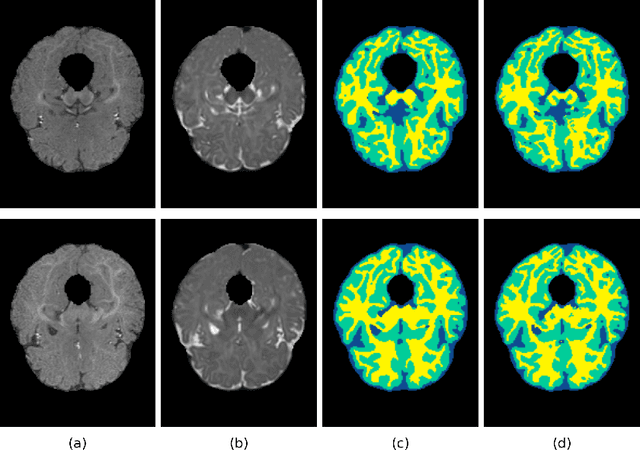
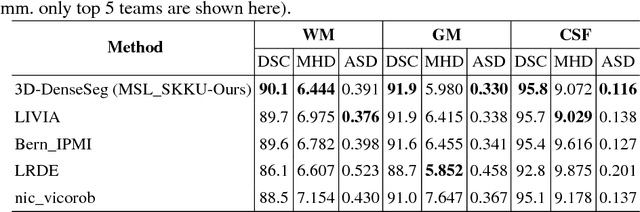
Abstract:In the isointense stage, the accurate volumetric image segmentation is a challenging task due to the low contrast between tissues. In this paper, we propose a novel very deep network architecture based on a densely convolutional network for volumetric brain segmentation. The proposed network architecture provides a dense connection between layers that aims to improve the information flow in the network. By concatenating features map of fine and coarse dense blocks, it allows capturing multi-scale contextual information. Experimental results demonstrate significant advantages of the proposed method over existing methods, in terms of both segmentation accuracy and parameter efficiency in MICCAI grand challenge on 6-month infant brain MRI segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge