Jitae Shin
Senior Member, IEEE
Uncertainty Measurement of Deep Learning System based on the Convex Hull of Training Sets
May 25, 2024Abstract:Deep Learning (DL) has made remarkable achievements in computer vision and adopted in safety critical domains such as medical imaging or autonomous drive. Thus, it is necessary to understand the uncertainty of the model to effectively reduce accidents and losses due to misjudgment of the Deep Neural Networks (DNN). This can start by efficiently selecting data that could potentially malfunction to the model. Traditionally, data collection and labeling have been done manually, but recently test data selection methods have emerged that focus on capturing samples that are not relevant to what the model had been learned. They're selected based on the activation pattern of neurons in DNN, entropy minimization based on softmax output of the DL. However, these methods cannot quantitatively analyze the extent to which unseen samples are extrapolated from the training data. Therefore, we propose To-hull Uncertainty and Closure Ratio, which measures an uncertainty of trained model based on the convex hull of training data. It can observe the positional relation between the convex hull of the learned data and an unseen sample and infer how extrapolate the sample is from the convex hull. To evaluate the proposed method, we conduct empirical studies on popular datasets and DNN models, compared to state-of-the art test selection metrics. As a result of the experiment, the proposed To-hull Uncertainty is effective in finding samples with unusual patterns (e.g. adversarial attack) compared to the existing test selection metric.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020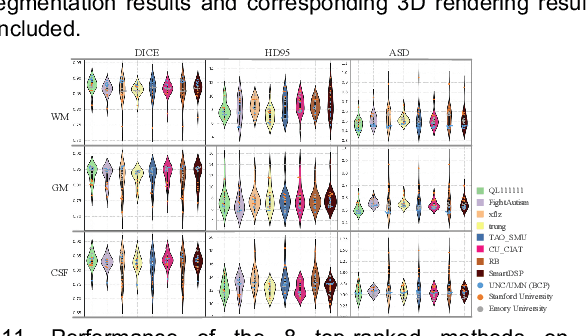
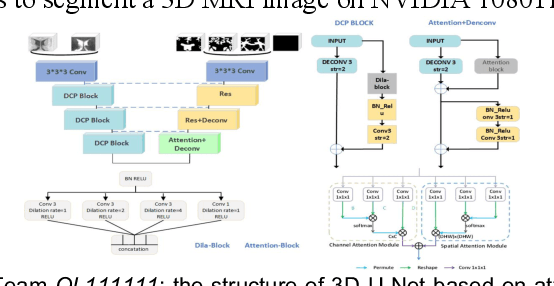
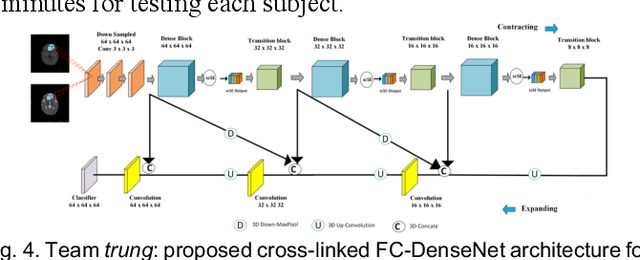
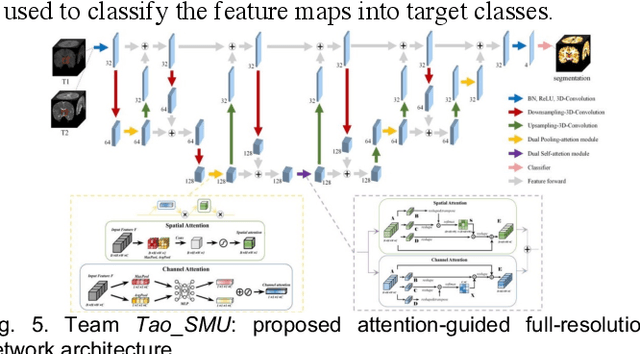
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
3D Densely Convolutional Networks for Volumetric Segmentation
Sep 13, 2017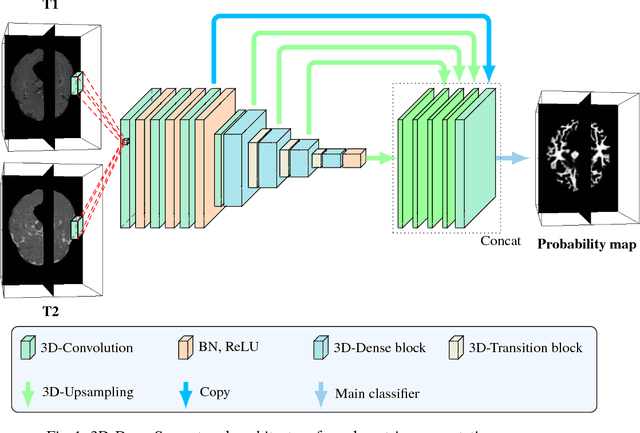
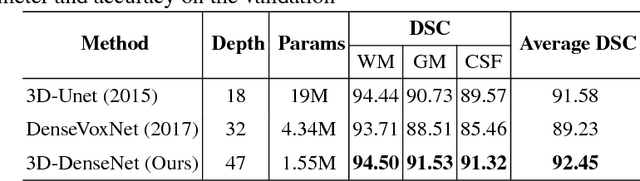
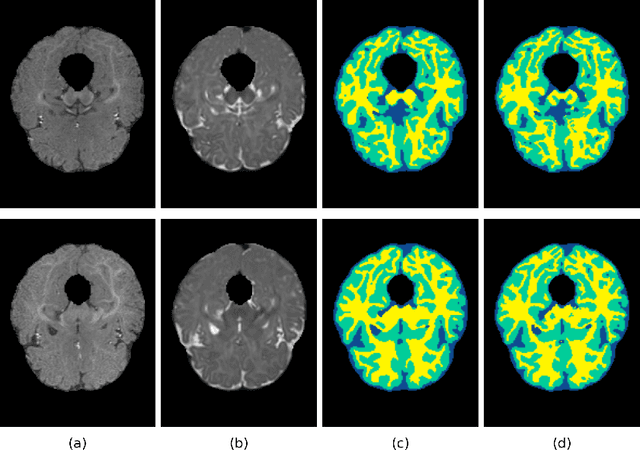
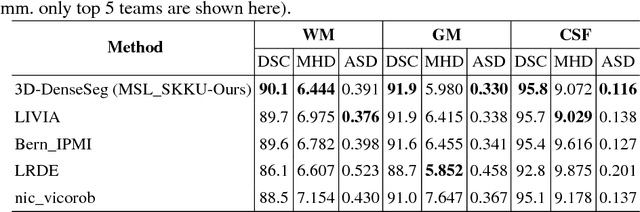
Abstract:In the isointense stage, the accurate volumetric image segmentation is a challenging task due to the low contrast between tissues. In this paper, we propose a novel very deep network architecture based on a densely convolutional network for volumetric brain segmentation. The proposed network architecture provides a dense connection between layers that aims to improve the information flow in the network. By concatenating features map of fine and coarse dense blocks, it allows capturing multi-scale contextual information. Experimental results demonstrate significant advantages of the proposed method over existing methods, in terms of both segmentation accuracy and parameter efficiency in MICCAI grand challenge on 6-month infant brain MRI segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge