Scott Mayer McKinney
From Medprompt to o1: Exploration of Run-Time Strategies for Medical Challenge Problems and Beyond
Nov 06, 2024



Abstract:Run-time steering strategies like Medprompt are valuable for guiding large language models (LLMs) to top performance on challenging tasks. Medprompt demonstrates that a general LLM can be focused to deliver state-of-the-art performance on specialized domains like medicine by using a prompt to elicit a run-time strategy involving chain of thought reasoning and ensembling. OpenAI's o1-preview model represents a new paradigm, where a model is designed to do run-time reasoning before generating final responses. We seek to understand the behavior of o1-preview on a diverse set of medical challenge problem benchmarks. Following on the Medprompt study with GPT-4, we systematically evaluate the o1-preview model across various medical benchmarks. Notably, even without prompting techniques, o1-preview largely outperforms the GPT-4 series with Medprompt. We further systematically study the efficacy of classic prompt engineering strategies, as represented by Medprompt, within the new paradigm of reasoning models. We found that few-shot prompting hinders o1's performance, suggesting that in-context learning may no longer be an effective steering approach for reasoning-native models. While ensembling remains viable, it is resource-intensive and requires careful cost-performance optimization. Our cost and accuracy analysis across run-time strategies reveals a Pareto frontier, with GPT-4o representing a more affordable option and o1-preview achieving state-of-the-art performance at higher cost. Although o1-preview offers top performance, GPT-4o with steering strategies like Medprompt retains value in specific contexts. Moreover, we note that the o1-preview model has reached near-saturation on many existing medical benchmarks, underscoring the need for new, challenging benchmarks. We close with reflections on general directions for inference-time computation with LLMs.
Can Generalist Foundation Models Outcompete Special-Purpose Tuning? Case Study in Medicine
Nov 28, 2023



Abstract:Generalist foundation models such as GPT-4 have displayed surprising capabilities in a wide variety of domains and tasks. Yet, there is a prevalent assumption that they cannot match specialist capabilities of fine-tuned models. For example, most explorations to date on medical competency benchmarks have leveraged domain-specific training, as exemplified by efforts on BioGPT and Med-PaLM. We build on a prior study of GPT-4's capabilities on medical challenge benchmarks in the absence of special training. Rather than using simple prompting to highlight the model's out-of-the-box capabilities, we perform a systematic exploration of prompt engineering. We find that prompting innovation can unlock deeper specialist capabilities and show that GPT-4 easily tops prior leading results for medical benchmarks. The prompting methods we explore are general purpose, and make no specific use of domain expertise, removing the need for expert-curated content. Our experimental design carefully controls for overfitting during the prompt engineering process. We introduce Medprompt, based on a composition of several prompting strategies. With Medprompt, GPT-4 achieves state-of-the-art results on all nine of the benchmark datasets in the MultiMedQA suite. The method outperforms leading specialist models such as Med-PaLM 2 by a significant margin with an order of magnitude fewer calls to the model. Steering GPT-4 with Medprompt achieves a 27% reduction in error rate on the MedQA dataset over the best methods to date achieved with specialist models and surpasses a score of 90% for the first time. Beyond medical problems, we show the power of Medprompt to generalize to other domains and provide evidence for the broad applicability of the approach via studies of the strategy on exams in electrical engineering, machine learning, philosophy, accounting, law, nursing, and clinical psychology.
Capabilities of GPT-4 on Medical Challenge Problems
Mar 20, 2023



Abstract:Large language models (LLMs) have demonstrated remarkable capabilities in natural language understanding and generation across various domains, including medicine. We present a comprehensive evaluation of GPT-4, a state-of-the-art LLM, on medical competency examinations and benchmark datasets. GPT-4 is a general-purpose model that is not specialized for medical problems through training or engineered to solve clinical tasks. Our analysis covers two sets of official practice materials for the USMLE, a three-step examination program used to assess clinical competency and grant licensure in the United States. We also evaluate performance on the MultiMedQA suite of benchmark datasets. Beyond measuring model performance, experiments were conducted to investigate the influence of test questions containing both text and images on model performance, probe for memorization of content during training, and study probability calibration, which is of critical importance in high-stakes applications like medicine. Our results show that GPT-4, without any specialized prompt crafting, exceeds the passing score on USMLE by over 20 points and outperforms earlier general-purpose models (GPT-3.5) as well as models specifically fine-tuned on medical knowledge (Med-PaLM, a prompt-tuned version of Flan-PaLM 540B). In addition, GPT-4 is significantly better calibrated than GPT-3.5, demonstrating a much-improved ability to predict the likelihood that its answers are correct. We also explore the behavior of the model qualitatively through a case study that shows the ability of GPT-4 to explain medical reasoning, personalize explanations to students, and interactively craft new counterfactual scenarios around a medical case. Implications of the findings are discussed for potential uses of GPT-4 in medical education, assessment, and clinical practice, with appropriate attention to challenges of accuracy and safety.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022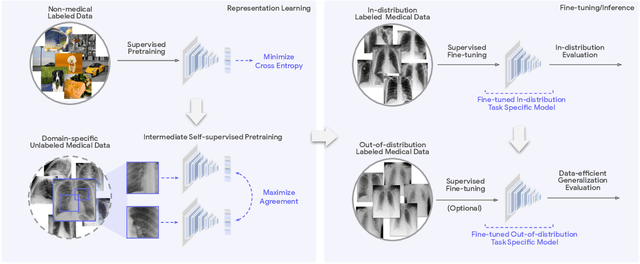
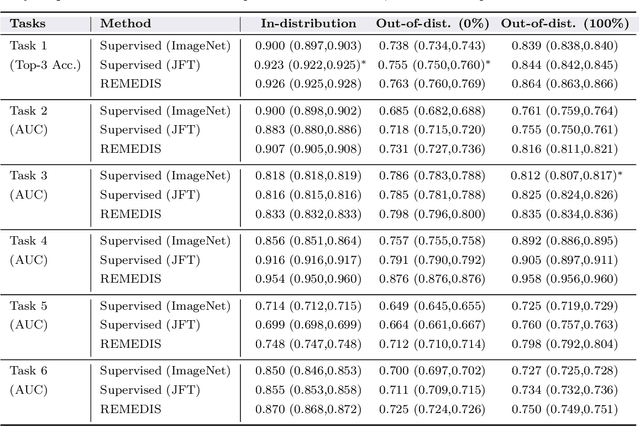
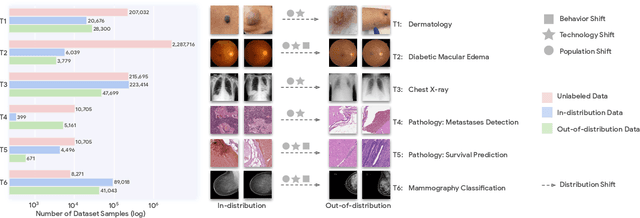
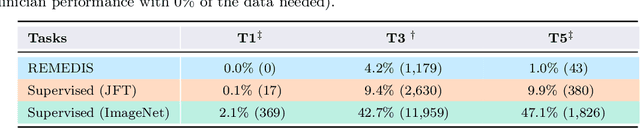
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
AI system for fetal ultrasound in low-resource settings
Mar 18, 2022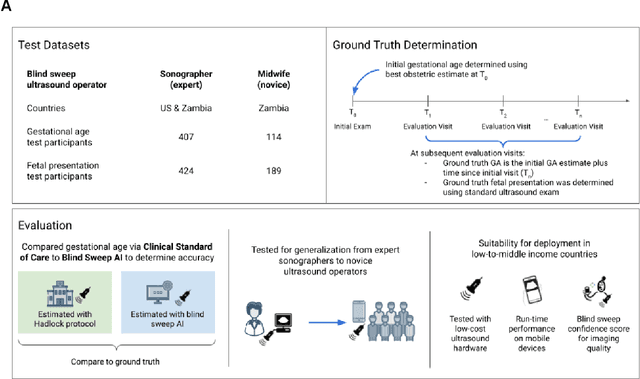
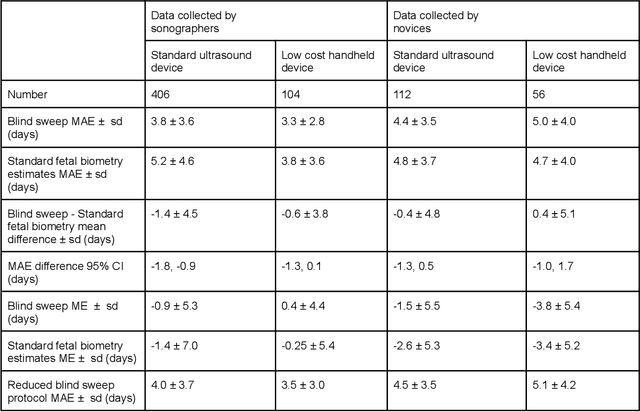
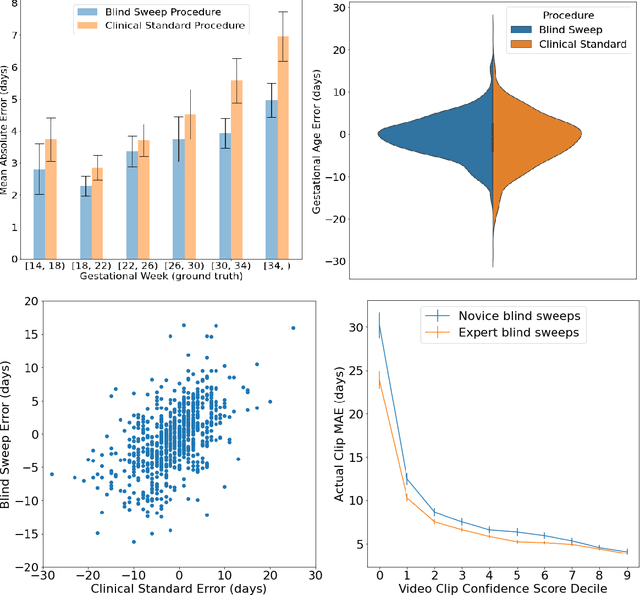
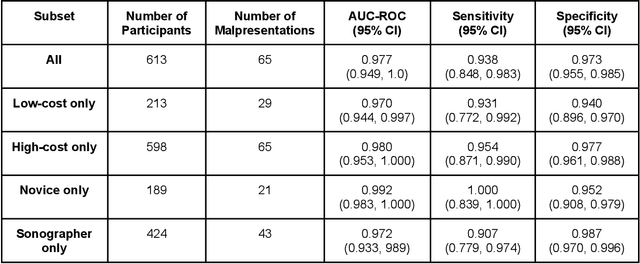
Abstract:Despite considerable progress in maternal healthcare, maternal and perinatal deaths remain high in low-to-middle income countries. Fetal ultrasound is an important component of antenatal care, but shortage of adequately trained healthcare workers has limited its adoption. We developed and validated an artificial intelligence (AI) system that uses novice-acquired "blind sweep" ultrasound videos to estimate gestational age (GA) and fetal malpresentation. We further addressed obstacles that may be encountered in low-resourced settings. Using a simplified sweep protocol with real-time AI feedback on sweep quality, we have demonstrated the generalization of model performance to minimally trained novice ultrasound operators using low cost ultrasound devices with on-device AI integration. The GA model was non-inferior to standard fetal biometry estimates with as few as two sweeps, and the fetal malpresentation model had high AUC-ROCs across operators and devices. Our AI models have the potential to assist in upleveling the capabilities of lightly trained ultrasound operators in low resource settings.
Deep learning for detecting pulmonary tuberculosis via chest radiography: an international study across 10 countries
May 16, 2021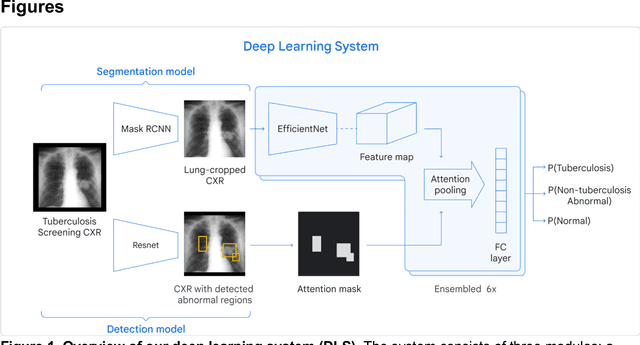
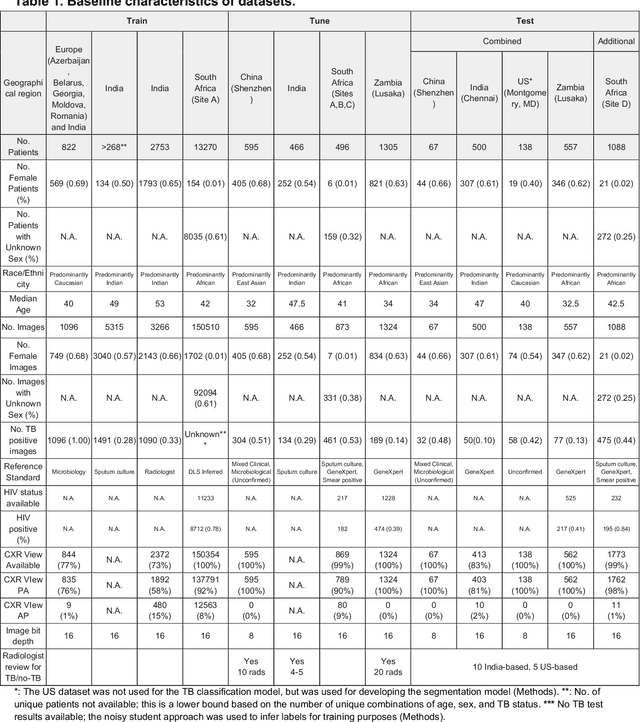
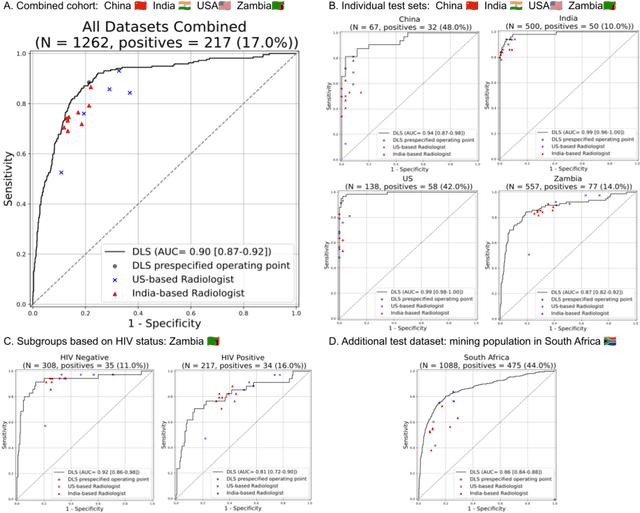
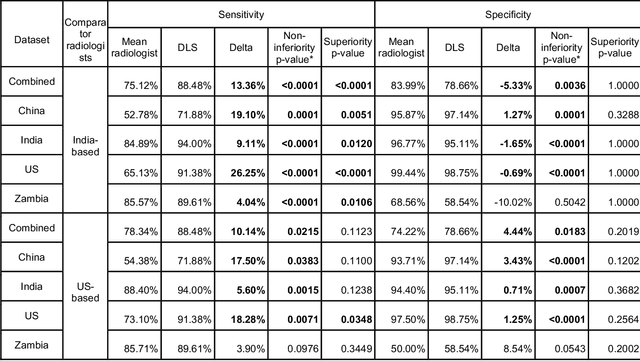
Abstract:Tuberculosis (TB) is a top-10 cause of death worldwide. Though the WHO recommends chest radiographs (CXRs) for TB screening, the limited availability of CXR interpretation is a barrier. We trained a deep learning system (DLS) to detect active pulmonary TB using CXRs from 9 countries across Africa, Asia, and Europe, and utilized large-scale CXR pretraining, attention pooling, and noisy student semi-supervised learning. Evaluation was on (1) a combined test set spanning China, India, US, and Zambia, and (2) an independent mining population in South Africa. Given WHO targets of 90% sensitivity and 70% specificity, the DLS's operating point was prespecified to favor sensitivity over specificity. On the combined test set, the DLS's ROC curve was above all 9 India-based radiologists, with an AUC of 0.90 (95%CI 0.87-0.92). The DLS's sensitivity (88%) was higher than the India-based radiologists (75% mean sensitivity), p<0.001 for superiority; and its specificity (79%) was non-inferior to the radiologists (84% mean specificity), p=0.004. Similar trends were observed within HIV positive and sputum smear positive sub-groups, and in the South Africa test set. We found that 5 US-based radiologists (where TB isn't endemic) were more sensitive and less specific than the India-based radiologists (where TB is endemic). The DLS also remained non-inferior to the US-based radiologists. In simulations, using the DLS as a prioritization tool for confirmatory testing reduced the cost per positive case detected by 40-80% compared to using confirmatory testing alone. To conclude, our DLS generalized to 5 countries, and merits prospective evaluation to assist cost-effective screening efforts in radiologist-limited settings. Operating point flexibility may permit customization of the DLS to account for site-specific factors such as TB prevalence, demographics, clinical resources, and customary practice patterns.
Supervised Transfer Learning at Scale for Medical Imaging
Jan 21, 2021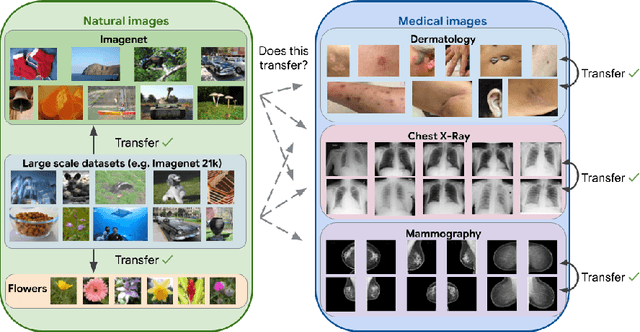
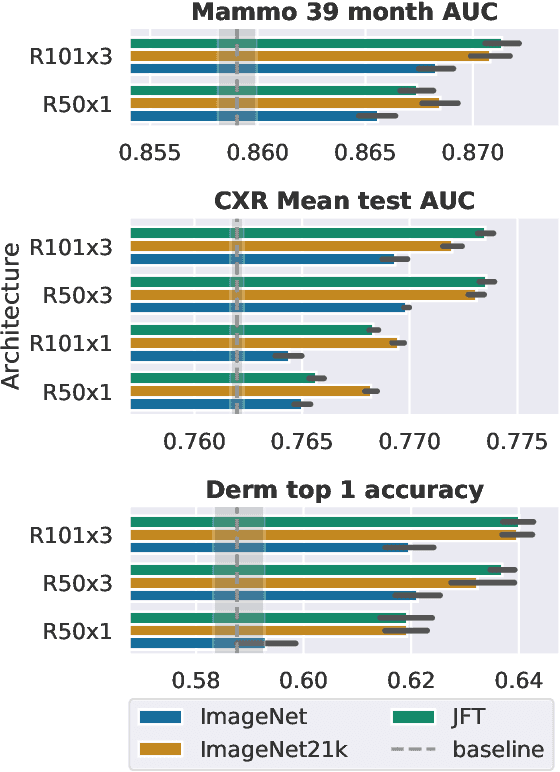
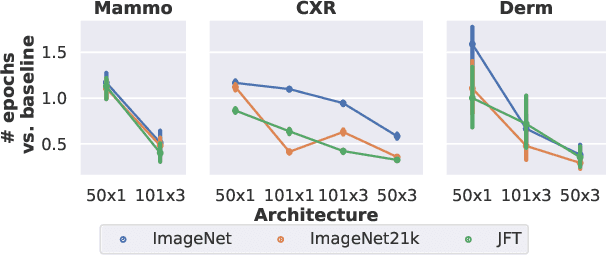
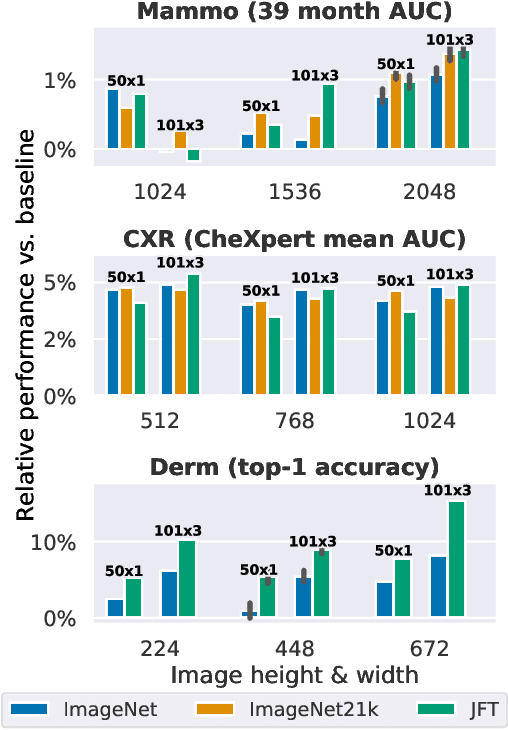
Abstract:Transfer learning is a standard technique to improve performance on tasks with limited data. However, for medical imaging, the value of transfer learning is less clear. This is likely due to the large domain mismatch between the usual natural-image pre-training (e.g. ImageNet) and medical images. However, recent advances in transfer learning have shown substantial improvements from scale. We investigate whether modern methods can change the fortune of transfer learning for medical imaging. For this, we study the class of large-scale pre-trained networks presented by Kolesnikov et al. on three diverse imaging tasks: chest radiography, mammography, and dermatology. We study both transfer performance and critical properties for the deployment in the medical domain, including: out-of-distribution generalization, data-efficiency, sub-group fairness, and uncertainty estimation. Interestingly, we find that for some of these properties transfer from natural to medical images is indeed extremely effective, but only when performed at sufficient scale.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge