Samuel Joutard
HyperSORT: Self-Organising Robust Training with hyper-networks
Jun 26, 2025Abstract:Medical imaging datasets often contain heterogeneous biases ranging from erroneous labels to inconsistent labeling styles. Such biases can negatively impact deep segmentation networks performance. Yet, the identification and characterization of such biases is a particularly tedious and challenging task. In this paper, we introduce HyperSORT, a framework using a hyper-network predicting UNets' parameters from latent vectors representing both the image and annotation variability. The hyper-network parameters and the latent vector collection corresponding to each data sample from the training set are jointly learned. Hence, instead of optimizing a single neural network to fit a dataset, HyperSORT learns a complex distribution of UNet parameters where low density areas can capture noise-specific patterns while larger modes robustly segment organs in differentiated but meaningful manners. We validate our method on two 3D abdominal CT public datasets: first a synthetically perturbed version of the AMOS dataset, and TotalSegmentator, a large scale dataset containing real unknown biases and errors. Our experiments show that HyperSORT creates a structured mapping of the dataset allowing the identification of relevant systematic biases and erroneous samples. Latent space clusters yield UNet parameters performing the segmentation task in accordance with the underlying learned systematic bias. The code and our analysis of the TotalSegmentator dataset are made available: https://github.com/ImFusionGmbH/HyperSORT
HyperSpace: Hypernetworks for spacing-adaptive image segmentation
Jul 04, 2024Abstract:Medical images are often acquired in different settings, requiring harmonization to adapt to the operating point of algorithms. Specifically, to standardize the physical spacing of imaging voxels in heterogeneous inference settings, images are typically resampled before being processed by deep learning models. However, down-sampling results in loss of information, whereas upsampling introduces redundant information leading to inefficient resource utilization. To overcome these issues, we propose to condition segmentation models on the voxel spacing using hypernetworks. Our approach allows processing images at their native resolutions or at resolutions adjusted to the hardware and time constraints at inference time. Our experiments across multiple datasets demonstrate that our approach achieves competitive performance compared to resolution-specific models, while offering greater flexibility for the end user. This also simplifies model development, deployment and maintenance. Our code is available at https://github.com/ImFusionGmbH/HyperSpace.
Unified Brain MR-Ultrasound Synthesis using Multi-Modal Hierarchical Representations
Sep 19, 2023Abstract:We introduce MHVAE, a deep hierarchical variational auto-encoder (VAE) that synthesizes missing images from various modalities. Extending multi-modal VAEs with a hierarchical latent structure, we introduce a probabilistic formulation for fusing multi-modal images in a common latent representation while having the flexibility to handle incomplete image sets as input. Moreover, adversarial learning is employed to generate sharper images. Extensive experiments are performed on the challenging problem of joint intra-operative ultrasound (iUS) and Magnetic Resonance (MR) synthesis. Our model outperformed multi-modal VAEs, conditional GANs, and the current state-of-the-art unified method (ResViT) for synthesizing missing images, demonstrating the advantage of using a hierarchical latent representation and a principled probabilistic fusion operation. Our code is publicly available \url{https://github.com/ReubenDo/MHVAE}.
Driving Points Prediction For Abdominal Probabilistic Registration
Aug 05, 2022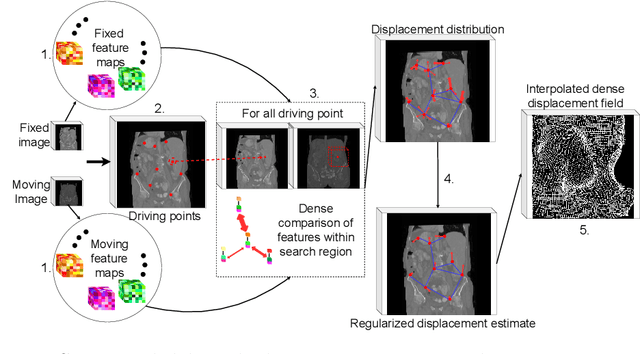
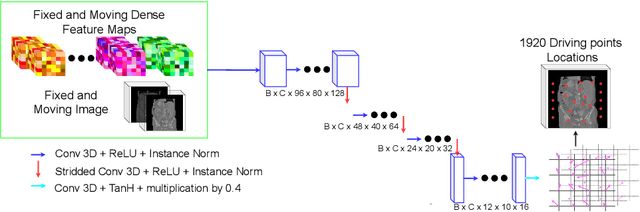
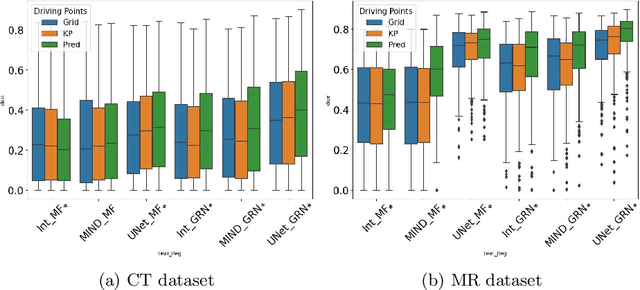
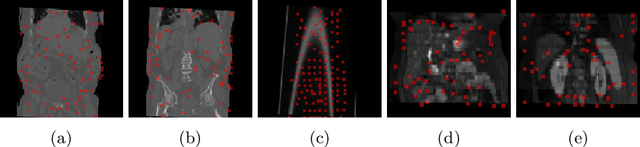
Abstract:Inter-patient abdominal registration has various applications, from pharmakinematic studies to anatomy modeling. Yet, it remains a challenging application due to the morphological heterogeneity and variability of the human abdomen. Among the various registration methods proposed for this task, probabilistic displacement registration models estimate displacement distribution for a subset of points by comparing feature vectors of points from the two images. These probabilistic models are informative and robust while allowing large displacements by design. As the displacement distributions are typically estimated on a subset of points (which we refer to as driving points), due to computational requirements, we propose in this work to learn a driving points predictor. Compared to previously proposed methods, the driving points predictor is optimized in an end-to-end fashion to infer driving points tailored for a specific registration pipeline. We evaluate the impact of our contribution on two different datasets corresponding to different modalities. Specifically, we compared the performances of 6 different probabilistic displacement registration models when using a driving points predictor or one of 2 other standard driving points selection methods. The proposed method improved performances in 11 out of 12 experiments.
A multi-organ point cloud registration algorithm for abdominal CT registration
Mar 15, 2022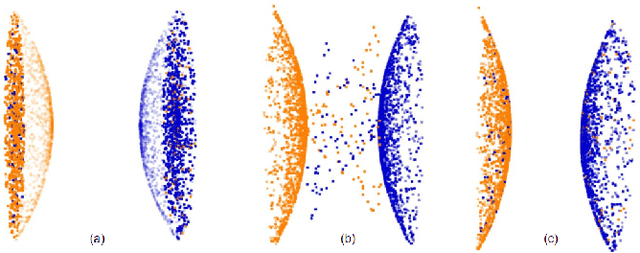
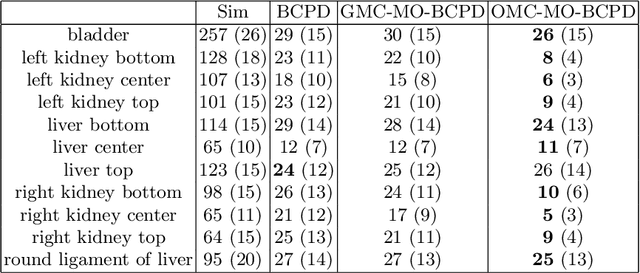
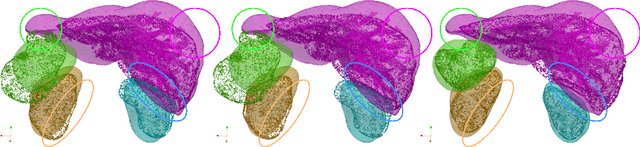
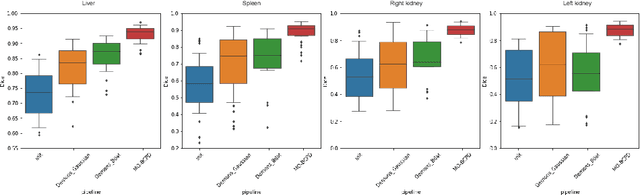
Abstract:Registering CT images of the chest is a crucial step for several tasks such as disease progression tracking or surgical planning. It is also a challenging step because of the heterogeneous content of the human abdomen which implies complex deformations. In this work, we focus on accurately registering a subset of organs of interest. We register organ surface point clouds, as may typically be extracted from an automatic segmentation pipeline, by expanding the Bayesian Coherent Point Drift algorithm (BCPD). We introduce MO-BCPD, a multi-organ version of the BCPD algorithm which explicitly models three important aspects of this task: organ individual elastic properties, inter-organ motion coherence and segmentation inaccuracy. This model also provides an interpolation framework to estimate the deformation of the entire volume. We demonstrate the efficiency of our method by registering different patients from the LITS challenge dataset. The target registration error on anatomical landmarks is almost twice as small for MO-BCPD compared to standard BCPD while imposing the same constraints on individual organs deformation.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022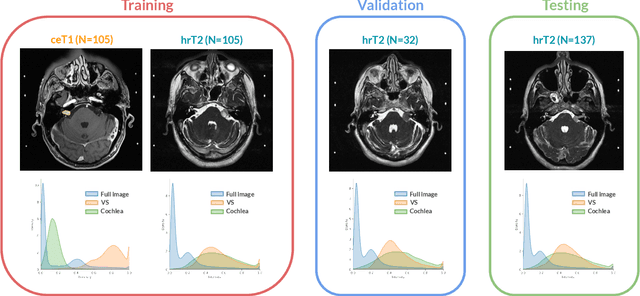
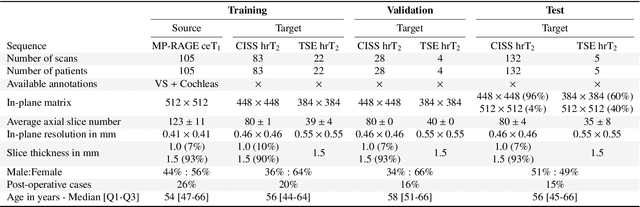
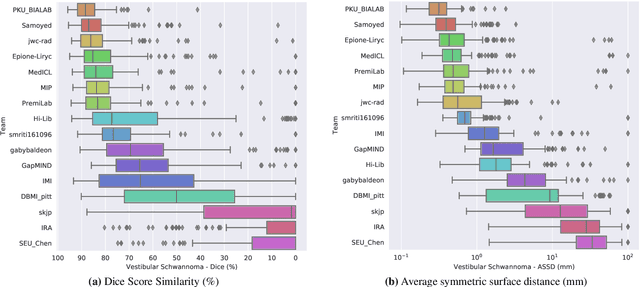
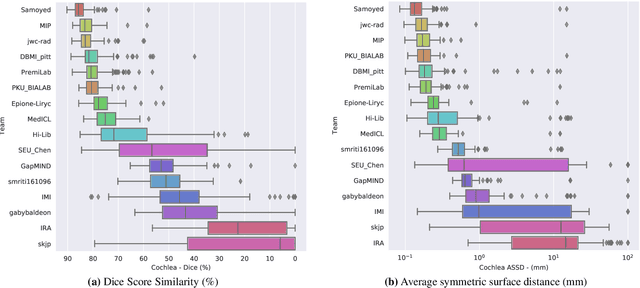
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
Inter Extreme Points Geodesics for Weakly Supervised Segmentation
Jul 01, 2021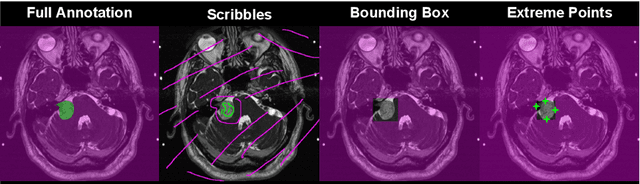
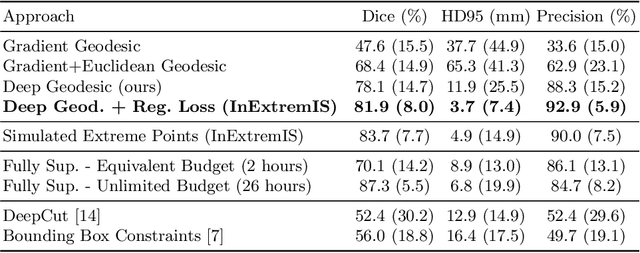
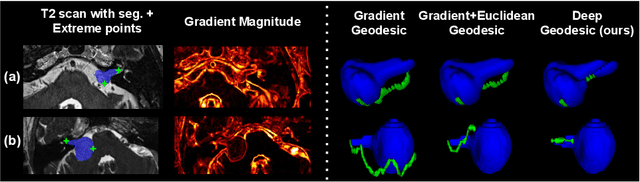
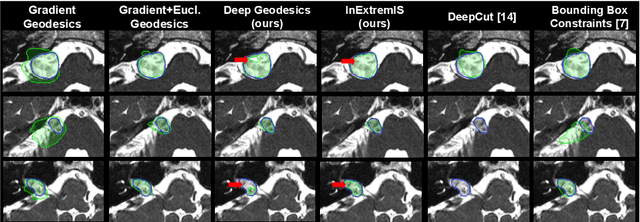
Abstract:We introduce $\textit{InExtremIS}$, a weakly supervised 3D approach to train a deep image segmentation network using particularly weak train-time annotations: only 6 extreme clicks at the boundary of the objects of interest. Our fully-automatic method is trained end-to-end and does not require any test-time annotations. From the extreme points, 3D bounding boxes are extracted around objects of interest. Then, deep geodesics connecting extreme points are generated to increase the amount of "annotated" voxels within the bounding boxes. Finally, a weakly supervised regularised loss derived from a Conditional Random Field formulation is used to encourage prediction consistency over homogeneous regions. Extensive experiments are performed on a large open dataset for Vestibular Schwannoma segmentation. $\textit{InExtremIS}$ obtained competitive performance, approaching full supervision and outperforming significantly other weakly supervised techniques based on bounding boxes. Moreover, given a fixed annotation time budget, $\textit{InExtremIS}$ outperforms full supervision. Our code and data are available online.
Scribble-based Domain Adaptation via Co-segmentation
Jul 07, 2020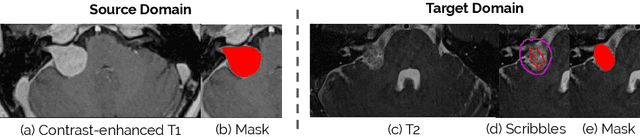
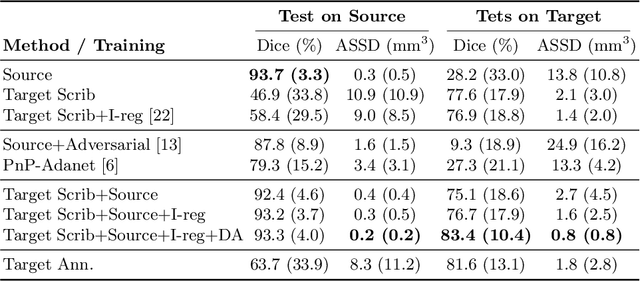
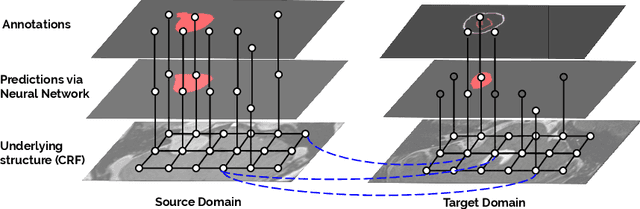
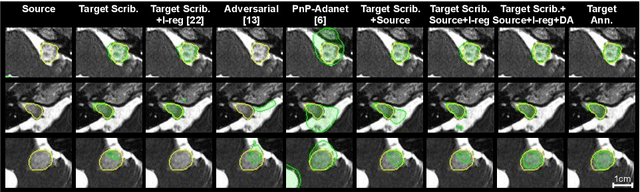
Abstract:Although deep convolutional networks have reached state-of-the-art performance in many medical image segmentation tasks, they have typically demonstrated poor generalisation capability. To be able to generalise from one domain (e.g. one imaging modality) to another, domain adaptation has to be performed. While supervised methods may lead to good performance, they require to fully annotate additional data which may not be an option in practice. In contrast, unsupervised methods don't need additional annotations but are usually unstable and hard to train. In this work, we propose a novel weakly-supervised method. Instead of requiring detailed but time-consuming annotations, scribbles on the target domain are used to perform domain adaptation. This paper introduces a new formulation of domain adaptation based on structured learning and co-segmentation. Our method is easy to train, thanks to the introduction of a regularised loss. The framework is validated on Vestibular Schwannoma segmentation (T1 to T2 scans). Our proposed method outperforms unsupervised approaches and achieves comparable performance to a fully-supervised approach.
Hetero-Modal Variational Encoder-Decoder for Joint Modality Completion and Segmentation
Jul 25, 2019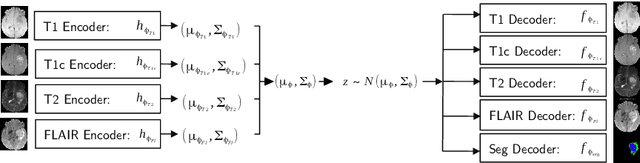

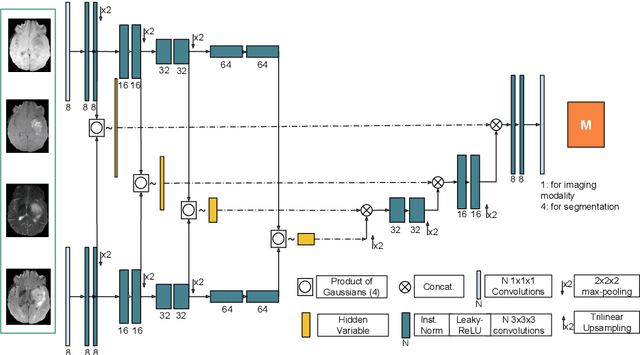
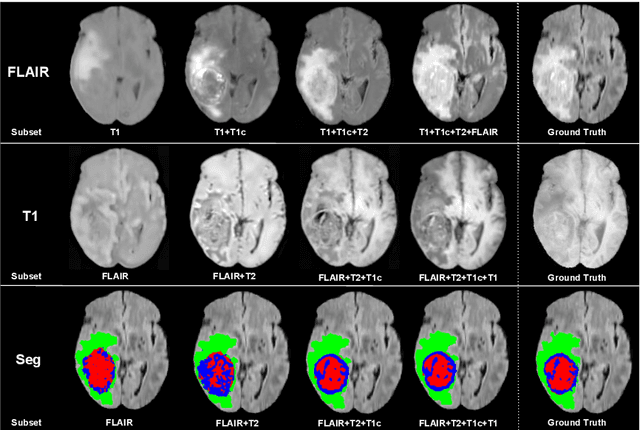
Abstract:We propose a new deep learning method for tumour segmentation when dealing with missing imaging modalities. Instead of producing one network for each possible subset of observed modalities or using arithmetic operations to combine feature maps, our hetero-modal variational 3D encoder-decoder independently embeds all observed modalities into a shared latent representation. Missing data and tumour segmentation can be then generated from this embedding. In our scenario, the input is a random subset of modalities. We demonstrate that the optimisation problem can be seen as a mixture sampling. In addition to this, we introduce a new network architecture building upon both the 3D U-Net and the Multi-Modal Variational Auto-Encoder (MVAE). Finally, we evaluate our method on BraTS2018 using subsets of the imaging modalities as input. Our model outperforms the current state-of-the-art method for dealing with missing modalities and achieves similar performance to the subset-specific equivalent networks.
Permutohedral Attention Module for Efficient Non-Local Neural Networks
Jul 01, 2019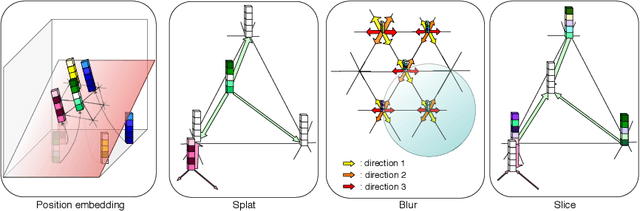
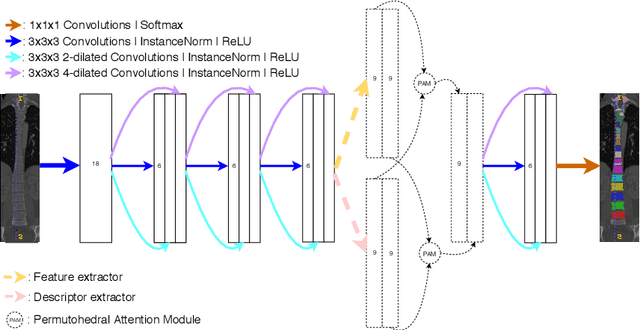
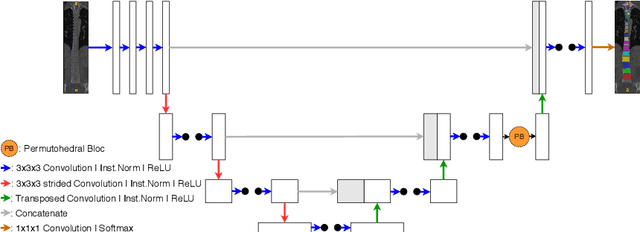
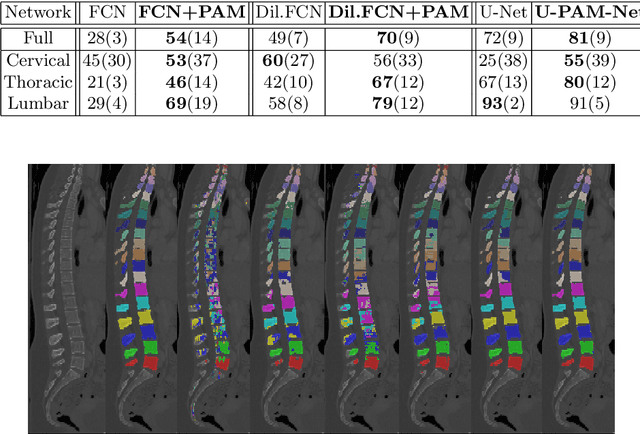
Abstract:Medical image processing tasks such as segmentation often require capturing non-local information. As organs, bones, and tissues share common characteristics such as intensity, shape, and texture, the contextual information plays a critical role in correctly labeling them. Segmentation and labeling is now typically done with convolutional neural networks (CNNs) but the context of the CNN is limited by the receptive field which itself is limited by memory requirements and other properties. In this paper, we propose a new attention module, that we call Permutohedral Attention Module (PAM), to efficiently capture non-local characteristics of the image. The proposed method is both memory and computationally efficient. We provide a GPU implementation of this module suitable for 3D medical imaging problems. We demonstrate the efficiency and scalability of our module with the challenging task of vertebrae segmentation and labeling where context plays a crucial role because of the very similar appearance of different vertebrae.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge