Kayhan Batmanghelich
A Cautionary Tale of Self-Supervised Learning for Imaging Biomarkers: Alzheimer's Disease Case Study
Jan 23, 2026Abstract:Discovery of sensitive and biologically grounded biomarkers is essential for early detection and monitoring of Alzheimer's disease (AD). Structural MRI is widely available but typically relies on hand-crafted features such as cortical thickness or volume. We ask whether self-supervised learning (SSL) can uncover more powerful biomarkers from the same data. Existing SSL methods underperform FreeSurfer-derived features in disease classification, conversion prediction, and amyloid status prediction. We introduce Residual Noise Contrastive Estimation (R-NCE), a new SSL framework that integrates auxiliary FreeSurfer features while maximizing additional augmentation-invariant information. R-NCE outperforms traditional features and existing SSL methods across multiple benchmarks, including AD conversion prediction. To assess biological relevance, we derive Brain Age Gap (BAG) measures and perform genome-wide association studies. R-NCE-BAG shows high heritability and associations with MAPT and IRAG1, with enrichment in astrocytes and oligodendrocytes, indicating sensitivity to neurodegenerative and cerebrovascular processes.
Semantic Consistency-Based Uncertainty Quantification for Factuality in Radiology Report Generation
Dec 05, 2024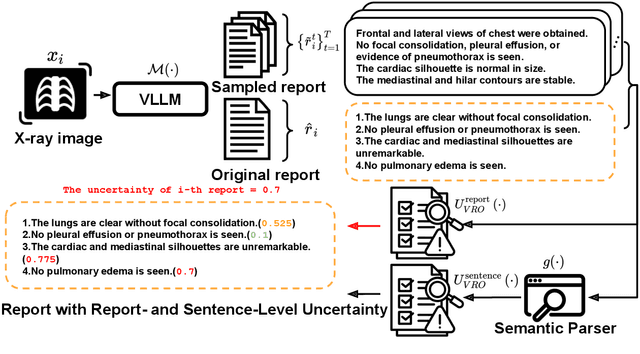
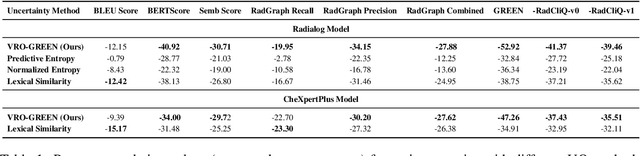
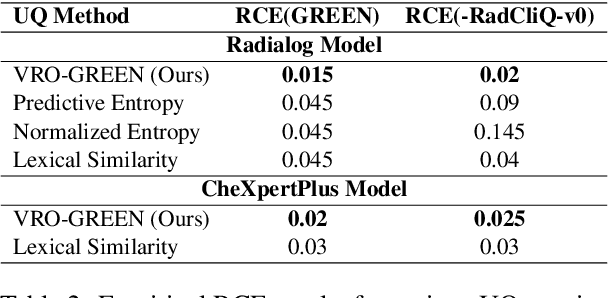
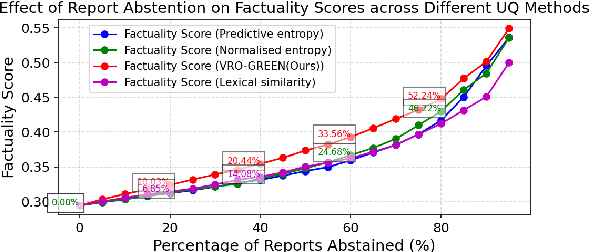
Abstract:Radiology report generation (RRG) has shown great potential in assisting radiologists by automating the labor-intensive task of report writing. While recent advancements have improved the quality and coherence of generated reports, ensuring their factual correctness remains a critical challenge. Although generative medical Vision Large Language Models (VLLMs) have been proposed to address this issue, these models are prone to hallucinations and can produce inaccurate diagnostic information. To address these concerns, we introduce a novel Semantic Consistency-Based Uncertainty Quantification framework that provides both report-level and sentence-level uncertainties. Unlike existing approaches, our method does not require modifications to the underlying model or access to its inner state, such as output token logits, thus serving as a plug-and-play module that can be seamlessly integrated with state-of-the-art models. Extensive experiments demonstrate the efficacy of our method in detecting hallucinations and enhancing the factual accuracy of automatically generated radiology reports. By abstaining from high-uncertainty reports, our approach improves factuality scores by $10$%, achieved by rejecting $20$% of reports using the Radialog model on the MIMIC-CXR dataset. Furthermore, sentence-level uncertainty flags the lowest-precision sentence in each report with an $82.9$% success rate.
Mammo-CLIP: A Vision Language Foundation Model to Enhance Data Efficiency and Robustness in Mammography
May 22, 2024Abstract:The lack of large and diverse training data on Computer-Aided Diagnosis (CAD) in breast cancer detection has been one of the concerns that impedes the adoption of the system. Recently, pre-training with large-scale image text datasets via Vision-Language models (VLM) (\eg CLIP) partially addresses the issue of robustness and data efficiency in computer vision (CV). This paper proposes Mammo-CLIP, the first VLM pre-trained on a substantial amount of screening mammogram-report pairs, addressing the challenges of dataset diversity and size. Our experiments on two public datasets demonstrate strong performance in classifying and localizing various mammographic attributes crucial for breast cancer detection, showcasing data efficiency and robustness similar to CLIP in CV. We also propose Mammo-FActOR, a novel feature attribution method, to provide spatial interpretation of representation with sentence-level granularity within mammography reports. Code is available publicly: \url{https://github.com/batmanlab/Mammo-CLIP}.
MedSyn: Text-guided Anatomy-aware Synthesis of High-Fidelity 3D CT Images
Oct 18, 2023



Abstract:This paper introduces an innovative methodology for producing high-quality 3D lung CT images guided by textual information. While diffusion-based generative models are increasingly used in medical imaging, current state-of-the-art approaches are limited to low-resolution outputs and underutilize radiology reports' abundant information. The radiology reports can enhance the generation process by providing additional guidance and offering fine-grained control over the synthesis of images. Nevertheless, expanding text-guided generation to high-resolution 3D images poses significant memory and anatomical detail-preserving challenges. Addressing the memory issue, we introduce a hierarchical scheme that uses a modified UNet architecture. We start by synthesizing low-resolution images conditioned on the text, serving as a foundation for subsequent generators for complete volumetric data. To ensure the anatomical plausibility of the generated samples, we provide further guidance by generating vascular, airway, and lobular segmentation masks in conjunction with the CT images. The model demonstrates the capability to use textual input and segmentation tasks to generate synthesized images. The results of comparative assessments indicate that our approach exhibits superior performance compared to the most advanced models based on GAN and diffusion techniques, especially in accurately retaining crucial anatomical features such as fissure lines, airways, and vascular structures. This innovation introduces novel possibilities. This study focuses on two main objectives: (1) the development of a method for creating images based on textual prompts and anatomical components, and (2) the capability to generate new images conditioning on anatomical elements. The advancements in image generation can be applied to enhance numerous downstream tasks.
Two-Step Active Learning for Instance Segmentation with Uncertainty and Diversity Sampling
Sep 28, 2023


Abstract:Training high-quality instance segmentation models requires an abundance of labeled images with instance masks and classifications, which is often expensive to procure. Active learning addresses this challenge by striving for optimum performance with minimal labeling cost by selecting the most informative and representative images for labeling. Despite its potential, active learning has been less explored in instance segmentation compared to other tasks like image classification, which require less labeling. In this study, we propose a post-hoc active learning algorithm that integrates uncertainty-based sampling with diversity-based sampling. Our proposed algorithm is not only simple and easy to implement, but it also delivers superior performance on various datasets. Its practical application is demonstrated on a real-world overhead imagery dataset, where it increases the labeling efficiency fivefold.
Dividing and Conquering a BlackBox to a Mixture of Interpretable Models: Route, Interpret, Repeat
Jul 12, 2023Abstract:ML model design either starts with an interpretable model or a Blackbox and explains it post hoc. Blackbox models are flexible but difficult to explain, while interpretable models are inherently explainable. Yet, interpretable models require extensive ML knowledge and tend to be less flexible and underperforming than their Blackbox variants. This paper aims to blur the distinction between a post hoc explanation of a Blackbox and constructing interpretable models. Beginning with a Blackbox, we iteratively carve out a mixture of interpretable experts (MoIE) and a residual network. Each interpretable model specializes in a subset of samples and explains them using First Order Logic (FOL), providing basic reasoning on concepts from the Blackbox. We route the remaining samples through a flexible residual. We repeat the method on the residual network until all the interpretable models explain the desired proportion of data. Our extensive experiments show that our route, interpret, and repeat approach (1) identifies a diverse set of instance-specific concepts with high concept completeness via MoIE without compromising in performance, (2) identifies the relatively ``harder'' samples to explain via residuals, (3) outperforms the interpretable by-design models by significant margins during test-time interventions, and (4) fixes the shortcut learned by the original Blackbox. The code for MoIE is publicly available at: \url{https://github.com/batmanlab/ICML-2023-Route-interpret-repeat}
* appeared as v5 of arXiv:2302.10289 which was replaced in error, which drifted into a different work, accepted in ICML 2023
Exploring the Lottery Ticket Hypothesis with Explainability Methods: Insights into Sparse Network Performance
Jul 07, 2023Abstract:Discovering a high-performing sparse network within a massive neural network is advantageous for deploying them on devices with limited storage, such as mobile phones. Additionally, model explainability is essential to fostering trust in AI. The Lottery Ticket Hypothesis (LTH) finds a network within a deep network with comparable or superior performance to the original model. However, limited study has been conducted on the success or failure of LTH in terms of explainability. In this work, we examine why the performance of the pruned networks gradually increases or decreases. Using Grad-CAM and Post-hoc concept bottleneck models (PCBMs), respectively, we investigate the explainability of pruned networks in terms of pixels and high-level concepts. We perform extensive experiments across vision and medical imaging datasets. As more weights are pruned, the performance of the network degrades. The discovered concepts and pixels from the pruned networks are inconsistent with the original network -- a possible reason for the drop in performance.
Distilling BlackBox to Interpretable models for Efficient Transfer Learning
Jun 10, 2023Abstract:Building generalizable AI models is one of the primary challenges in the healthcare domain. While radiologists rely on generalizable descriptive rules of abnormality, Neural Network (NN) models suffer even with a slight shift in input distribution (e.g., scanner type). Fine-tuning a model to transfer knowledge from one domain to another requires a significant amount of labeled data in the target domain. In this paper, we develop an interpretable model that can be efficiently fine-tuned to an unseen target domain with minimal computational cost. We assume the interpretable component of NN to be approximately domain-invariant. However, interpretable models typically underperform compared to their Blackbox (BB) variants. We start with a BB in the source domain and distill it into a \emph{mixture} of shallow interpretable models using human-understandable concepts. As each interpretable model covers a subset of data, a mixture of interpretable models achieves comparable performance as BB. Further, we use the pseudo-labeling technique from semi-supervised learning (SSL) to learn the concept classifier in the target domain, followed by fine-tuning the interpretable models in the target domain. We evaluate our model using a real-life large-scale chest-X-ray (CXR) classification dataset. The code is available at: \url{https://github.com/batmanlab/MICCAI-2023-Route-interpret-repeat-CXRs}.
From Characters to Words: Hierarchical Pre-trained Language Model for Open-vocabulary Language Understanding
May 23, 2023Abstract:Current state-of-the-art models for natural language understanding require a preprocessing step to convert raw text into discrete tokens. This process known as tokenization relies on a pre-built vocabulary of words or sub-word morphemes. This fixed vocabulary limits the model's robustness to spelling errors and its capacity to adapt to new domains. In this work, we introduce a novel open-vocabulary language model that adopts a hierarchical two-level approach: one at the word level and another at the sequence level. Concretely, we design an intra-word module that uses a shallow Transformer architecture to learn word representations from their characters, and a deep inter-word Transformer module that contextualizes each word representation by attending to the entire word sequence. Our model thus directly operates on character sequences with explicit awareness of word boundaries, but without biased sub-word or word-level vocabulary. Experiments on various downstream tasks show that our method outperforms strong baselines. We also demonstrate that our hierarchical model is robust to textual corruption and domain shift.
DrasCLR: A Self-supervised Framework of Learning Disease-related and Anatomy-specific Representation for 3D Medical Images
Mar 15, 2023



Abstract:Large-scale volumetric medical images with annotation are rare, costly, and time prohibitive to acquire. Self-supervised learning (SSL) offers a promising pre-training and feature extraction solution for many downstream tasks, as it only uses unlabeled data. Recently, SSL methods based on instance discrimination have gained popularity in the medical imaging domain. However, SSL pre-trained encoders may use many clues in the image to discriminate an instance that are not necessarily disease-related. Moreover, pathological patterns are often subtle and heterogeneous, requiring the ability of the desired method to represent anatomy-specific features that are sensitive to abnormal changes in different body parts. In this work, we present a novel SSL framework, named DrasCLR, for 3D medical imaging to overcome these challenges. We propose two domain-specific contrastive learning strategies: one aims to capture subtle disease patterns inside a local anatomical region, and the other aims to represent severe disease patterns that span larger regions. We formulate the encoder using conditional hyper-parameterized network, in which the parameters are dependant on the anatomical location, to extract anatomically sensitive features. Extensive experiments on large-scale computer tomography (CT) datasets of lung images show that our method improves the performance of many downstream prediction and segmentation tasks. The patient-level representation improves the performance of the patient survival prediction task. We show how our method can detect emphysema subtypes via dense prediction. We demonstrate that fine-tuning the pre-trained model can significantly reduce annotation efforts without sacrificing emphysema detection accuracy. Our ablation study highlights the importance of incorporating anatomical context into the SSL framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge