Pranav Rajpurkar
Harvard University
Evaluating Contextual Intelligence in Recyclability: A Comprehensive Study of Image-Based Reasoning Systems
Dec 31, 2025Abstract:While the importance of efficient recycling is widely acknowledged, accurately determining the recyclability of items and their proper disposal remains a complex task for the general public. In this study, we explore the application of cutting-edge vision-language models (GPT-4o, GPT-4o-mini, and Claude 3.5) for predicting the recyclability of commonly disposed items. Utilizing a curated dataset of images, we evaluated the models' ability to match objects to appropriate recycling bins, including assessing whether the items could physically fit into the available bins. Additionally, we investigated the models' performance across several challenging scenarios: (i) adjusting predictions based on location-specific recycling guidelines; (ii) accounting for contamination or structural damage; and (iii) handling objects composed of multiple materials. Our findings highlight the significant advancements in contextual understanding offered by these models compared to previous iterations, while also identifying areas where they still fall short. The continued refinement of context-aware models is crucial for enhancing public recycling practices and advancing environmental sustainability.
ReX-MLE: The Autonomous Agent Benchmark for Medical Imaging Challenges
Dec 19, 2025



Abstract:Autonomous coding agents built on large language models (LLMs) can now solve many general software and machine learning tasks, but they remain ineffective on complex, domain-specific scientific problems. Medical imaging is a particularly demanding domain, requiring long training cycles, high-dimensional data handling, and specialized preprocessing and validation pipelines, capabilities not fully measured in existing agent benchmarks. To address this gap, we introduce ReX-MLE, a benchmark of 20 challenges derived from high-impact medical imaging competitions spanning diverse modalities and task types. Unlike prior ML-agent benchmarks, ReX-MLE evaluates full end-to-end workflows, requiring agents to independently manage data preprocessing, model training, and submission under realistic compute and time constraints. Evaluating state-of-the-art agents (AIDE, ML-Master, R&D-Agent) with different LLM backends (GPT-5, Gemini, Claude), we observe a severe performance gap: most submissions rank in the 0th percentile compared to human experts. Failures stem from domain-knowledge and engineering limitations. ReX-MLE exposes these bottlenecks and provides a foundation for developing domain-aware autonomous AI systems.
RadGame: An AI-Powered Platform for Radiology Education
Sep 16, 2025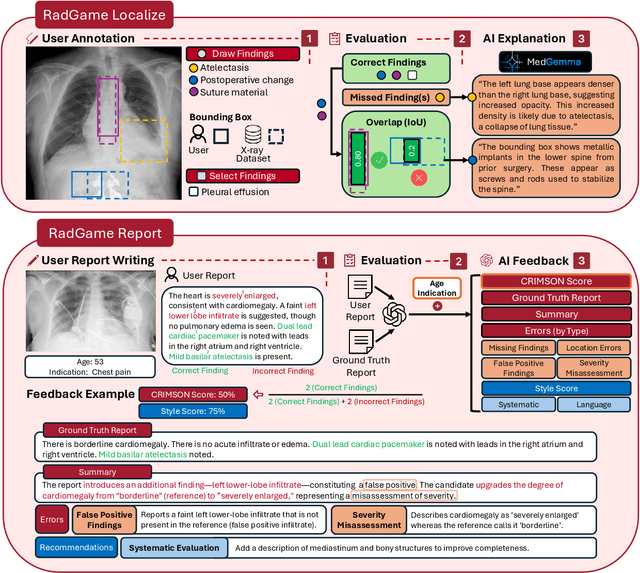
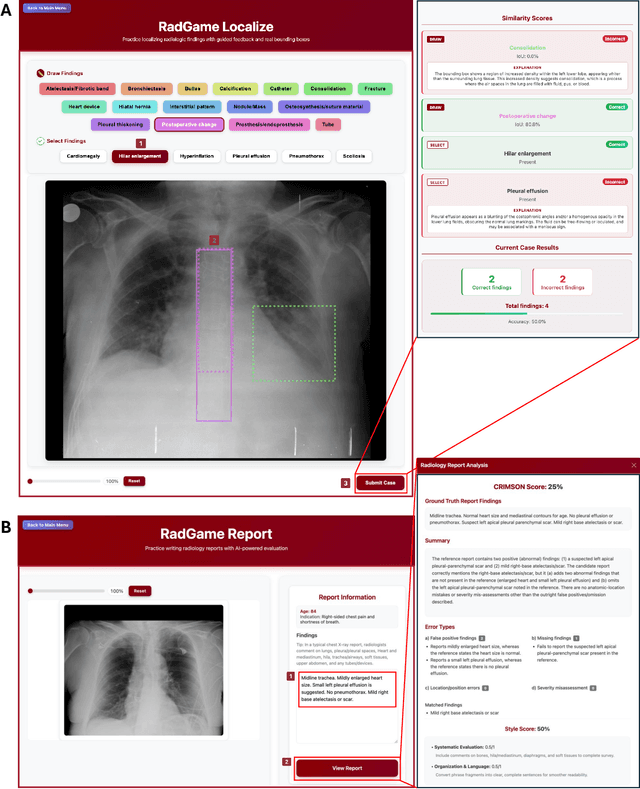
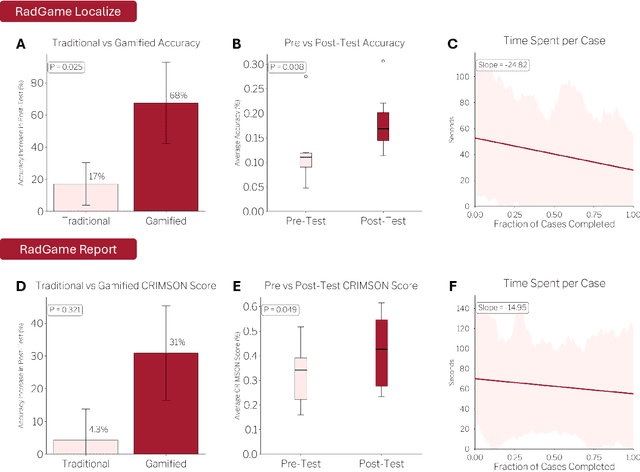
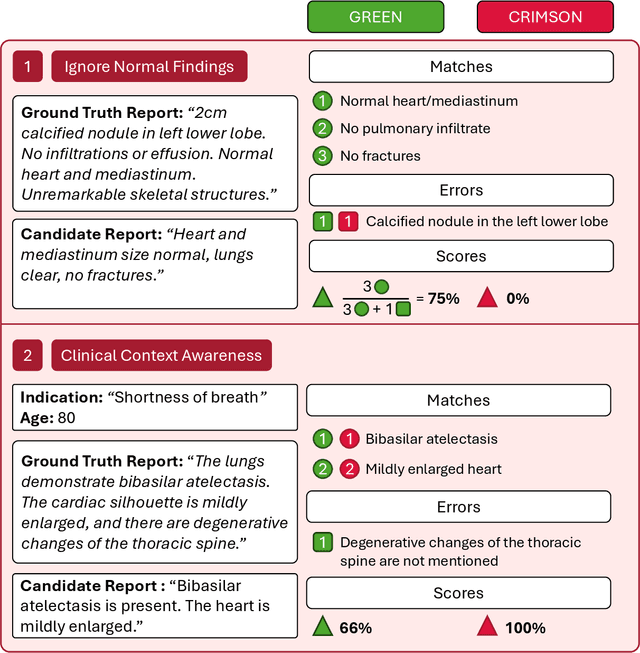
Abstract:We introduce RadGame, an AI-powered gamified platform for radiology education that targets two core skills: localizing findings and generating reports. Traditional radiology training is based on passive exposure to cases or active practice with real-time input from supervising radiologists, limiting opportunities for immediate and scalable feedback. RadGame addresses this gap by combining gamification with large-scale public datasets and automated, AI-driven feedback that provides clear, structured guidance to human learners. In RadGame Localize, players draw bounding boxes around abnormalities, which are automatically compared to radiologist-drawn annotations from public datasets, and visual explanations are generated by vision-language models for user missed findings. In RadGame Report, players compose findings given a chest X-ray, patient age and indication, and receive structured AI feedback based on radiology report generation metrics, highlighting errors and omissions compared to a radiologist's written ground truth report from public datasets, producing a final performance and style score. In a prospective evaluation, participants using RadGame achieved a 68% improvement in localization accuracy compared to 17% with traditional passive methods and a 31% improvement in report-writing accuracy compared to 4% with traditional methods after seeing the same cases. RadGame highlights the potential of AI-driven gamification to deliver scalable, feedback-rich radiology training and reimagines the application of medical AI resources in education.
ReXGroundingCT: A 3D Chest CT Dataset for Segmentation of Findings from Free-Text Reports
Jul 29, 2025Abstract:We present ReXGroundingCT, the first publicly available dataset to link free-text radiology findings with pixel-level segmentations in 3D chest CT scans that is manually annotated. While prior datasets have relied on structured labels or predefined categories, ReXGroundingCT captures the full expressiveness of clinical language represented in free text and grounds it to spatially localized 3D segmentation annotations in volumetric imaging. This addresses a critical gap in medical AI: the ability to connect complex, descriptive text, such as "3 mm nodule in the left lower lobe", to its precise anatomical location in three-dimensional space, a capability essential for grounded radiology report generation systems. The dataset comprises 3,142 non-contrast chest CT scans paired with standardized radiology reports from the CT-RATE dataset. Using a systematic three-stage pipeline, GPT-4 was used to extract positive lung and pleural findings, which were then manually segmented by expert annotators. A total of 8,028 findings across 16,301 entities were annotated, with quality control performed by board-certified radiologists. Approximately 79% of findings are focal abnormalities, while 21% are non-focal. The training set includes up to three representative segmentations per finding, while the validation and test sets contain exhaustive labels for each finding entity. ReXGroundingCT establishes a new benchmark for developing and evaluating sentence-level grounding and free-text medical segmentation models in chest CT. The dataset can be accessed at https://huggingface.co/datasets/rajpurkarlab/ReXGroundingCT.
ReXVQA: A Large-scale Visual Question Answering Benchmark for Generalist Chest X-ray Understanding
Jun 04, 2025



Abstract:We present ReXVQA, the largest and most comprehensive benchmark for visual question answering (VQA) in chest radiology, comprising approximately 696,000 questions paired with 160,000 chest X-rays studies across training, validation, and test sets. Unlike prior efforts that rely heavily on template based queries, ReXVQA introduces a diverse and clinically authentic task suite reflecting five core radiological reasoning skills: presence assessment, location analysis, negation detection, differential diagnosis, and geometric reasoning. We evaluate eight state-of-the-art multimodal large language models, including MedGemma-4B-it, Qwen2.5-VL, Janus-Pro-7B, and Eagle2-9B. The best-performing model (MedGemma) achieves 83.24% overall accuracy. To bridge the gap between AI performance and clinical expertise, we conducted a comprehensive human reader study involving 3 radiology residents on 200 randomly sampled cases. Our evaluation demonstrates that MedGemma achieved superior performance (83.84% accuracy) compared to human readers (best radiology resident: 77.27%), representing a significant milestone where AI performance exceeds expert human evaluation on chest X-ray interpretation. The reader study reveals distinct performance patterns between AI models and human experts, with strong inter-reader agreement among radiologists while showing more variable agreement patterns between human readers and AI models. ReXVQA establishes a new standard for evaluating generalist radiological AI systems, offering public leaderboards, fine-grained evaluation splits, structured explanations, and category-level breakdowns. This benchmark lays the foundation for next-generation AI systems capable of mimicking expert-level clinical reasoning beyond narrow pathology classification. Our dataset will be open-sourced at https://huggingface.co/datasets/rajpurkarlab/ReXVQA
ReXGradient-160K: A Large-Scale Publicly Available Dataset of Chest Radiographs with Free-text Reports
May 01, 2025Abstract:We present ReXGradient-160K, representing the largest publicly available chest X-ray dataset to date in terms of the number of patients. This dataset contains 160,000 chest X-ray studies with paired radiological reports from 109,487 unique patients across 3 U.S. health systems (79 medical sites). This comprehensive dataset includes multiple images per study and detailed radiology reports, making it particularly valuable for the development and evaluation of AI systems for medical imaging and automated report generation models. The dataset is divided into training (140,000 studies), validation (10,000 studies), and public test (10,000 studies) sets, with an additional private test set (10,000 studies) reserved for model evaluation on the ReXrank benchmark. By providing this extensive dataset, we aim to accelerate research in medical imaging AI and advance the state-of-the-art in automated radiological analysis. Our dataset will be open-sourced at https://huggingface.co/datasets/rajpurkarlab/ReXGradient-160K.
FreeTumor: Large-Scale Generative Tumor Synthesis in Computed Tomography Images for Improving Tumor Recognition
Feb 23, 2025Abstract:Tumor is a leading cause of death worldwide, with an estimated 10 million deaths attributed to tumor-related diseases every year. AI-driven tumor recognition unlocks new possibilities for more precise and intelligent tumor screening and diagnosis. However, the progress is heavily hampered by the scarcity of annotated datasets, which demands extensive annotation efforts by radiologists. To tackle this challenge, we introduce FreeTumor, an innovative Generative AI (GAI) framework to enable large-scale tumor synthesis for mitigating data scarcity. Specifically, FreeTumor effectively leverages a combination of limited labeled data and large-scale unlabeled data for tumor synthesis training. Unleashing the power of large-scale data, FreeTumor is capable of synthesizing a large number of realistic tumors on images for augmenting training datasets. To this end, we create the largest training dataset for tumor synthesis and recognition by curating 161,310 publicly available Computed Tomography (CT) volumes from 33 sources, with only 2.3% containing annotated tumors. To validate the fidelity of synthetic tumors, we engaged 13 board-certified radiologists in a Visual Turing Test to discern between synthetic and real tumors. Rigorous clinician evaluation validates the high quality of our synthetic tumors, as they achieved only 51.1% sensitivity and 60.8% accuracy in distinguishing our synthetic tumors from real ones. Through high-quality tumor synthesis, FreeTumor scales up the recognition training datasets by over 40 times, showcasing a notable superiority over state-of-the-art AI methods including various synthesis methods and foundation models. These findings indicate promising prospects of FreeTumor in clinical applications, potentially advancing tumor treatments and improving the survival rates of patients.
A Data-Efficient Pan-Tumor Foundation Model for Oncology CT Interpretation
Feb 10, 2025



Abstract:Artificial intelligence-assisted imaging analysis has made substantial strides in tumor diagnosis and management. Here we present PASTA, a pan-tumor CT foundation model that achieves state-of-the-art performance on 45 of 46 representative oncology tasks -- including lesion segmentation, tumor detection in plain CT, tumor staging, survival prediction, structured report generation, and cross-modality transfer learning, significantly outperforming the second-best models on 35 tasks. This remarkable advancement is driven by our development of PASTA-Gen, an innovative synthetic tumor generation framework that produces a comprehensive dataset of 30,000 CT scans with pixel-level annotated lesions and paired structured reports, encompassing malignancies across ten organs and five benign lesion types. By leveraging this rich, high-quality synthetic data, we overcome a longstanding bottleneck in the development of CT foundation models -- specifically, the scarcity of publicly available, high-quality annotated datasets due to privacy constraints and the substantial labor required for scaling precise data annotation. Encouragingly, PASTA demonstrates exceptional data efficiency with promising practical value, markedly improving performance on various tasks with only a small amount of real-world data. The open release of both the synthetic dataset and PASTA foundation model effectively addresses the challenge of data scarcity, thereby advancing oncological research and clinical translation.
a2z-1 for Multi-Disease Detection in Abdomen-Pelvis CT: External Validation and Performance Analysis Across 21 Conditions
Dec 17, 2024Abstract:We present a comprehensive evaluation of a2z-1, an artificial intelligence (AI) model designed to analyze abdomen-pelvis CT scans for 21 time-sensitive and actionable findings. Our study focuses on rigorous assessment of the model's performance and generalizability. Large-scale retrospective analysis demonstrates an average AUC of 0.931 across 21 conditions. External validation across two distinct health systems confirms consistent performance (AUC 0.923), establishing generalizability to different evaluation scenarios, with notable performance in critical findings such as small bowel obstruction (AUC 0.958) and acute pancreatitis (AUC 0.961). Subgroup analysis shows consistent accuracy across patient sex, age groups, and varied imaging protocols, including different slice thicknesses and contrast administration types. Comparison of high-confidence model outputs to radiologist reports reveals instances where a2z-1 identified overlooked findings, suggesting potential for quality assurance applications.
ReXTrust: A Model for Fine-Grained Hallucination Detection in AI-Generated Radiology Reports
Dec 17, 2024



Abstract:The increasing adoption of AI-generated radiology reports necessitates robust methods for detecting hallucinations--false or unfounded statements that could impact patient care. We present ReXTrust, a novel framework for fine-grained hallucination detection in AI-generated radiology reports. Our approach leverages sequences of hidden states from large vision-language models to produce finding-level hallucination risk scores. We evaluate ReXTrust on a subset of the MIMIC-CXR dataset and demonstrate superior performance compared to existing approaches, achieving an AUROC of 0.8751 across all findings and 0.8963 on clinically significant findings. Our results show that white-box approaches leveraging model hidden states can provide reliable hallucination detection for medical AI systems, potentially improving the safety and reliability of automated radiology reporting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge