Michael A. Riegler
Combining datasets to increase the number of samples and improve model fitting
Oct 11, 2022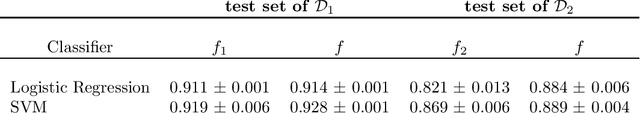

Abstract:For many use cases, combining information from different datasets can be of interest to improve a machine learning model's performance, especially when the number of samples from at least one of the datasets is small. However, a potential challenge in such cases is that the features from these datasets are not identical, even though there are some commonly shared features among the datasets. To tackle this challenge, we propose a novel framework called Combine datasets based on Imputation (ComImp). In addition, we propose a variant of ComImp that uses Principle Component Analysis (PCA), PCA-ComImp in order to reduce dimension before combining datasets. This is useful when the datasets have a large number of features that are not shared between them. Furthermore, our framework can also be utilized for data preprocessing by imputing missing data, i.e., filling in the missing entries while combining different datasets. To illustrate the power of the proposed methods and their potential usages, we conduct experiments for various tasks: regression, classification, and for different data types: tabular data, time series data, when the datasets to be combined have missing data. We also investigate how the devised methods can be used with transfer learning to provide even further model training improvement. Our results indicate that the proposed methods are somewhat similar to transfer learning in that the merge can significantly improve the accuracy of a prediction model on smaller datasets. In addition, the methods can boost performance by a significant margin when combining small datasets together and can provide extra improvement when being used with transfer learning.
Towards the Neuroevolution of Low-level Artificial General Intelligence
Jul 27, 2022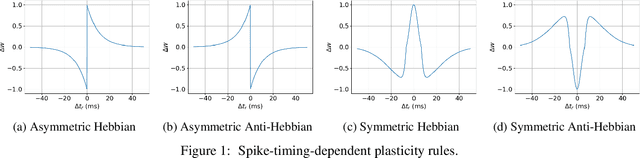
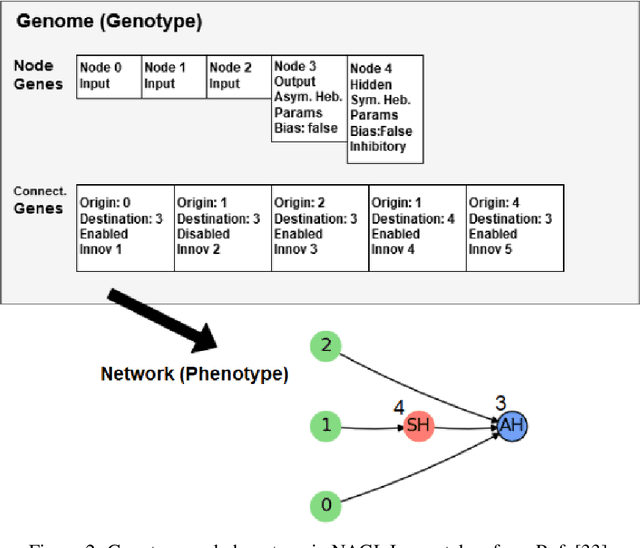
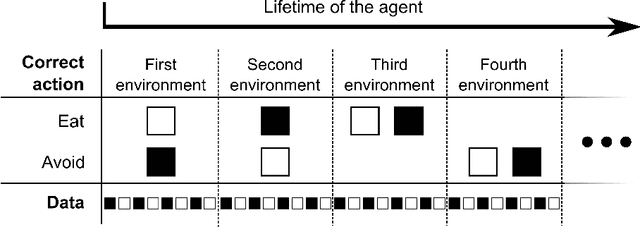
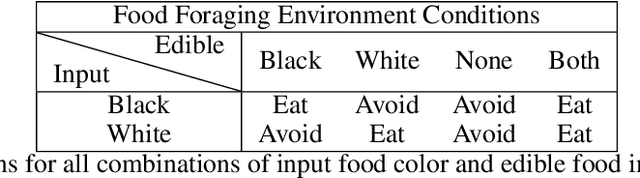
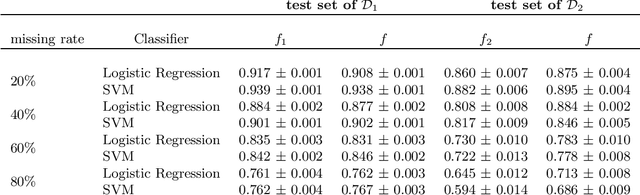
Abstract:In this work, we argue that the search for Artificial General Intelligence (AGI) should start from a much lower level than human-level intelligence. The circumstances of intelligent behavior in nature resulted from an organism interacting with its surrounding environment, which could change over time and exert pressure on the organism to allow for learning of new behaviors or environment models. Our hypothesis is that learning occurs through interpreting sensory feedback when an agent acts in an environment. For that to happen, a body and a reactive environment are needed. We evaluate a method to evolve a biologically-inspired artificial neural network that learns from environment reactions named Neuroevolution of Artificial General Intelligence (NAGI), a framework for low-level AGI. This method allows the evolutionary complexification of a randomly-initialized spiking neural network with adaptive synapses, which controls agents instantiated in mutable environments. Such a configuration allows us to benchmark the adaptivity and generality of the controllers. The chosen tasks in the mutable environments are food foraging, emulation of logic gates, and cart-pole balancing. The three tasks are successfully solved with rather small network topologies and therefore it opens up the possibility of experimenting with more complex tasks and scenarios where curriculum learning is beneficial.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022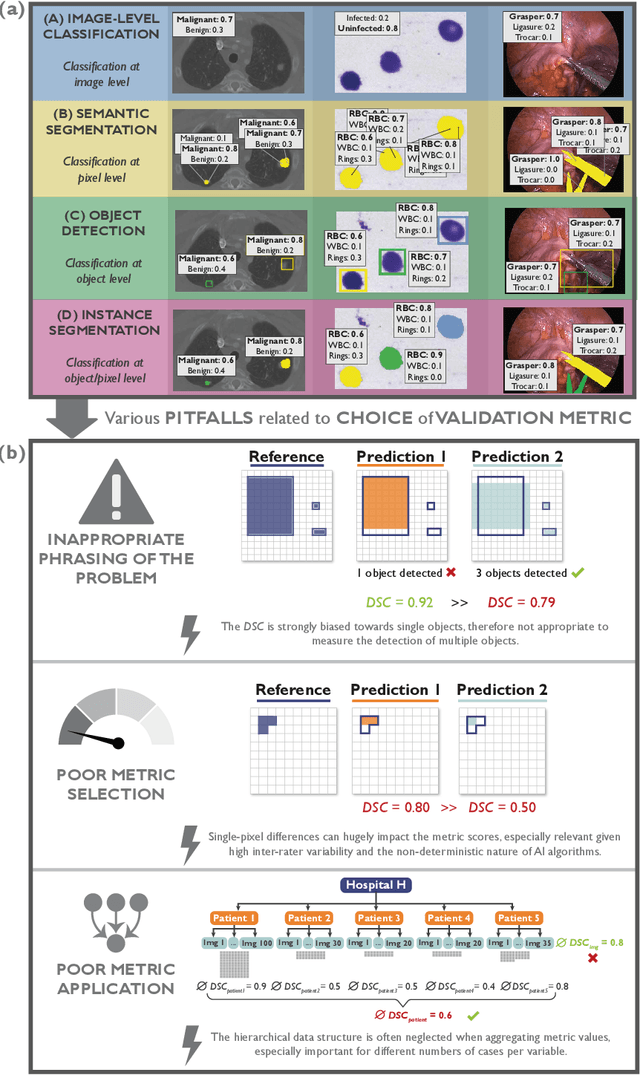
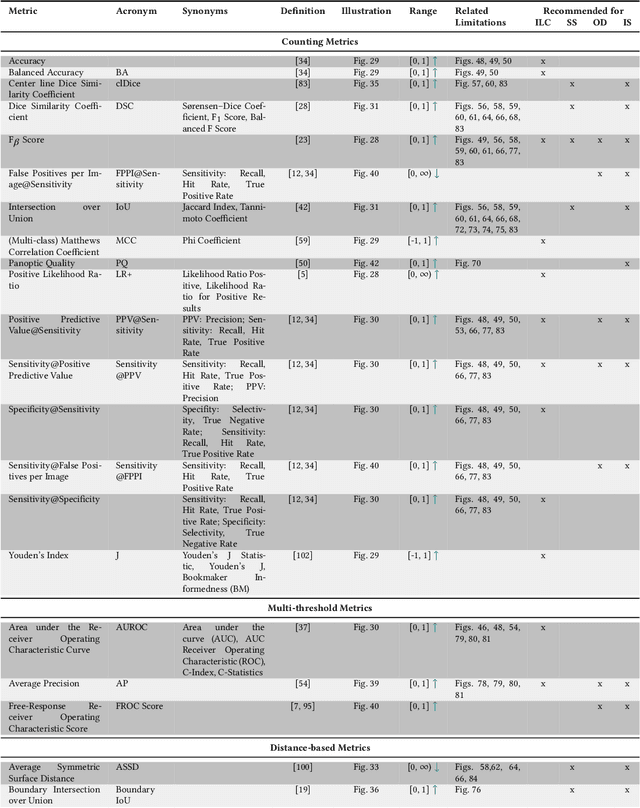
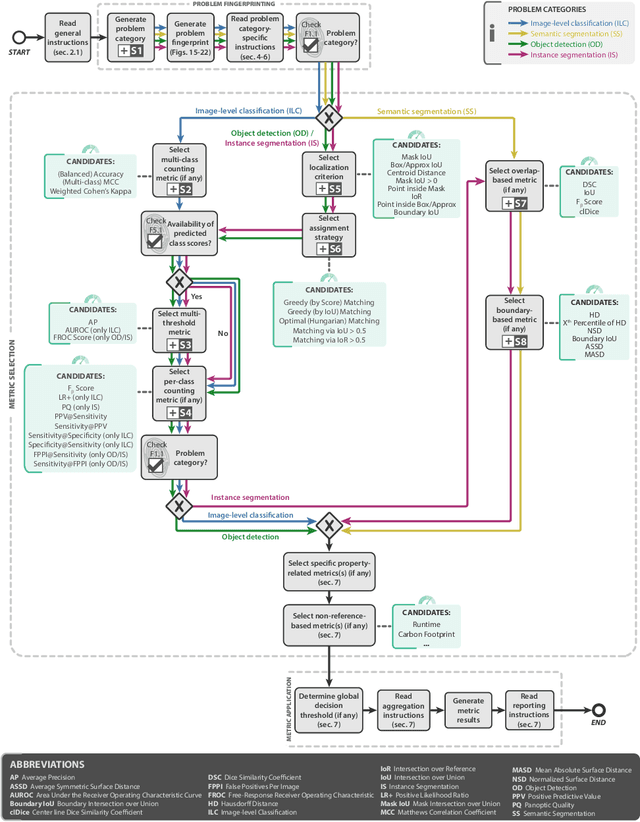
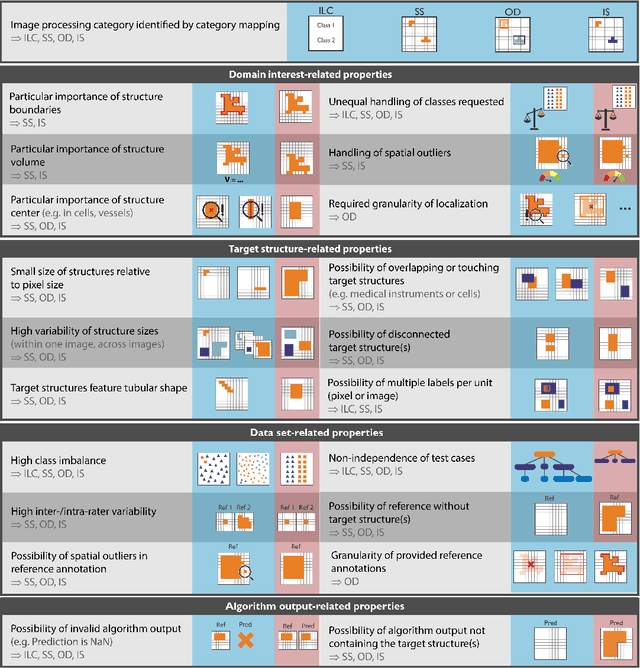
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
Segmentation Consistency Training: Out-of-Distribution Generalization for Medical Image Segmentation
May 30, 2022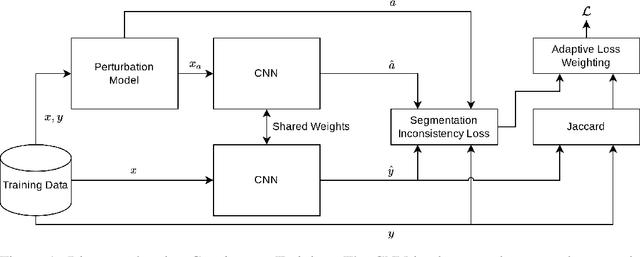

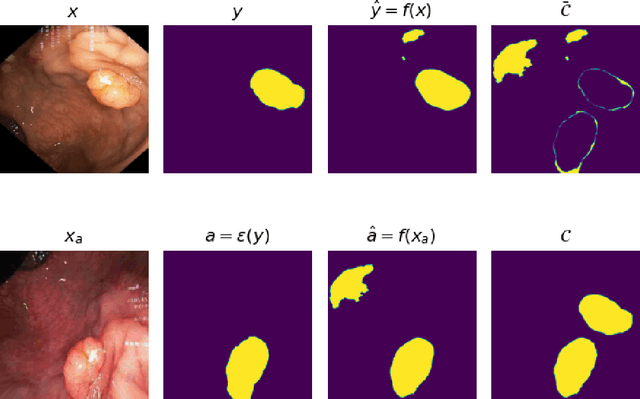
Abstract:Generalizability is seen as one of the major challenges in deep learning, in particular in the domain of medical imaging, where a change of hospital or in imaging routines can lead to a complete failure of a model. To tackle this, we introduce Consistency Training, a training procedure and alternative to data augmentation based on maximizing models' prediction consistency across augmented and unaugmented data in order to facilitate better out-of-distribution generalization. To this end, we develop a novel region-based segmentation loss function called Segmentation Inconsistency Loss (SIL), which considers the differences between pairs of augmented and unaugmented predictions and labels. We demonstrate that Consistency Training outperforms conventional data augmentation on several out-of-distribution datasets on polyp segmentation, a popular medical task.
PolypConnect: Image inpainting for generating realistic gastrointestinal tract images with polyps
May 30, 2022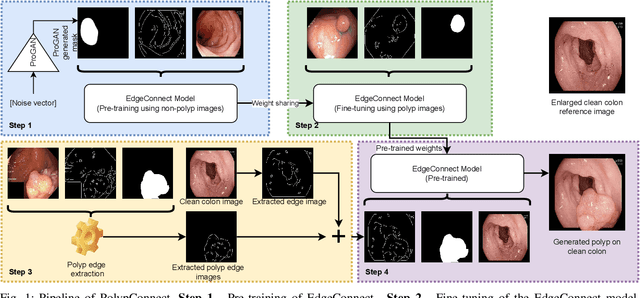
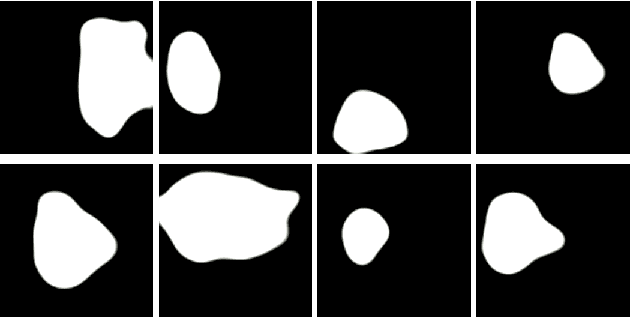

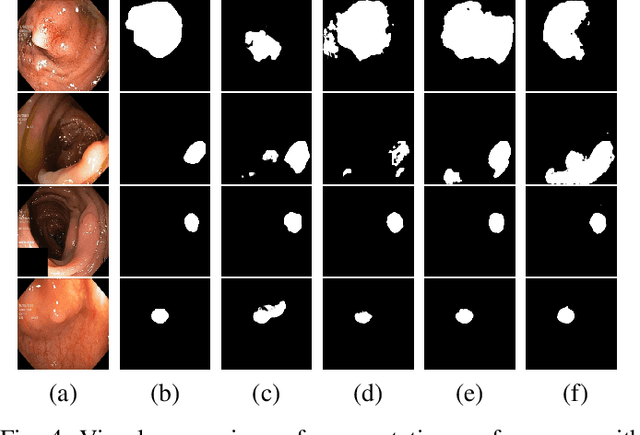
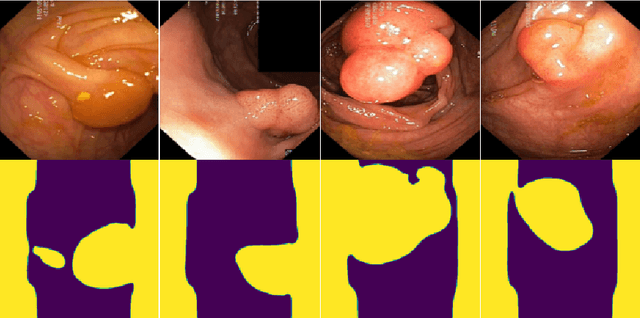
Abstract:Early identification of a polyp in the lower gastrointestinal (GI) tract can lead to prevention of life-threatening colorectal cancer. Developing computer-aided diagnosis (CAD) systems to detect polyps can improve detection accuracy and efficiency and save the time of the domain experts called endoscopists. Lack of annotated data is a common challenge when building CAD systems. Generating synthetic medical data is an active research area to overcome the problem of having relatively few true positive cases in the medical domain. To be able to efficiently train machine learning (ML) models, which are the core of CAD systems, a considerable amount of data should be used. In this respect, we propose the PolypConnect pipeline, which can convert non-polyp images into polyp images to increase the size of training datasets for training. We present the whole pipeline with quantitative and qualitative evaluations involving endoscopists. The polyp segmentation model trained using synthetic data, and real data shows a 5.1% improvement of mean intersection over union (mIOU), compared to the model trained only using real data. The codes of all the experiments are available on GitHub to reproduce the results.
Grid HTM: Hierarchical Temporal Memory for Anomaly Detection in Videos
May 30, 2022
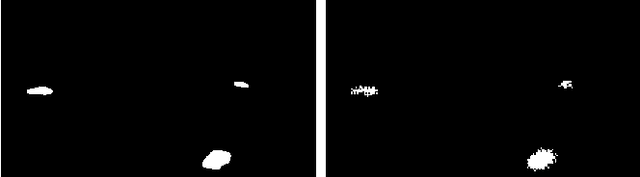
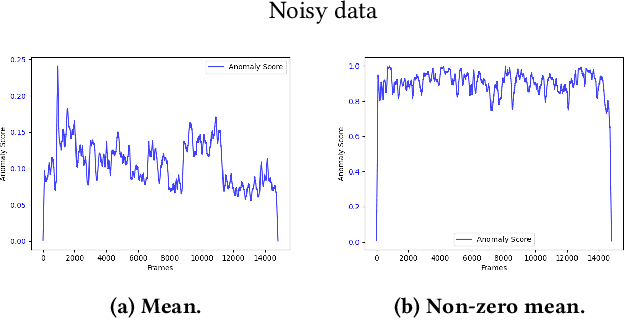
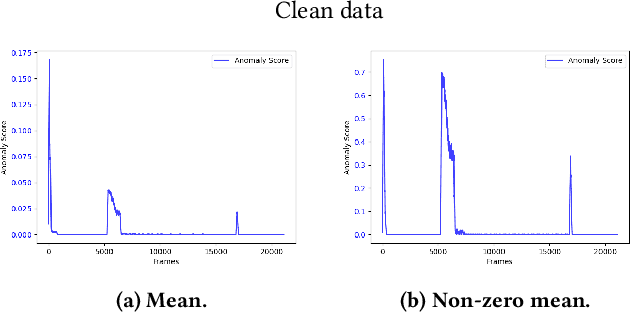
Abstract:The interest for video anomaly detection systems has gained traction for the past few years. The current approaches use deep learning to perform anomaly detection in videos, but this approach has multiple problems. For starters, deep learning in general has issues with noise, concept drift, explainability, and training data volumes. Additionally, anomaly detection in itself is a complex task and faces challenges such as unknowness, heterogeneity, and class imbalance. Anomaly detection using deep learning is therefore mainly constrained to generative models such as generative adversarial networks and autoencoders due to their unsupervised nature, but even they suffer from general deep learning issues and are hard to train properly. In this paper, we explore the capabilities of the Hierarchical Temporal Memory (HTM) algorithm to perform anomaly detection in videos, as it has favorable properties such as noise tolerance and online learning which combats concept drift. We introduce a novel version of HTM, namely, Grid HTM, which is an HTM-based architecture specifically for anomaly detection in complex videos such as surveillance footage.
Predicting tacrolimus exposure in kidney transplanted patients using machine learning
May 09, 2022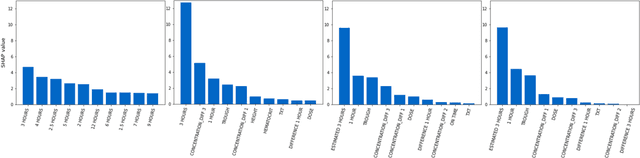
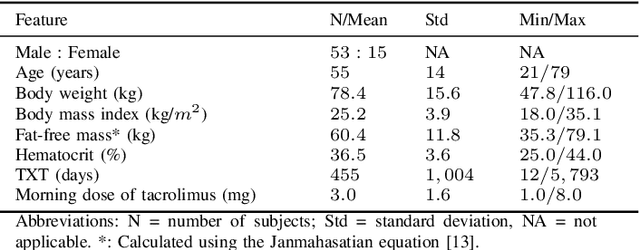
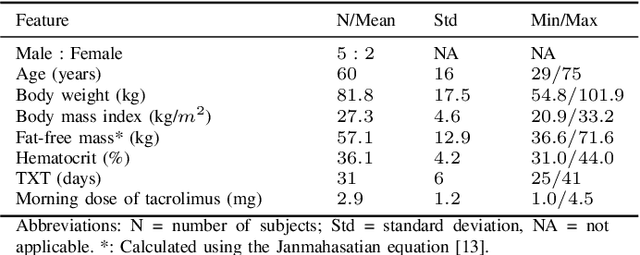
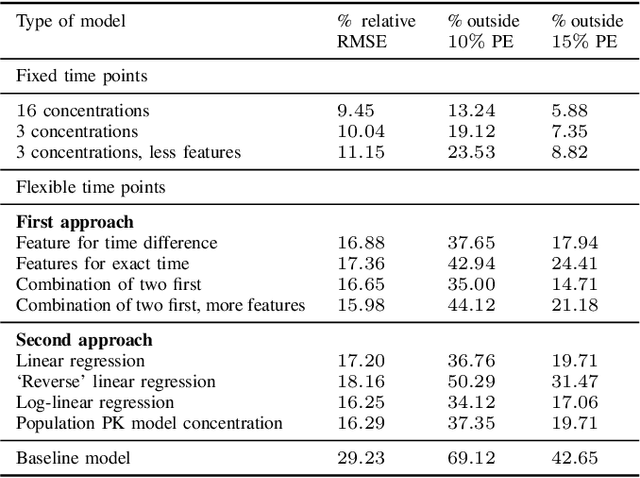
Abstract:Tacrolimus is one of the cornerstone immunosuppressive drugs in most transplantation centers worldwide following solid organ transplantation. Therapeutic drug monitoring of tacrolimus is necessary in order to avoid rejection of the transplanted organ or severe side effects. However, finding the right dose for a given patient is challenging, even for experienced clinicians. Consequently, a tool that can accurately estimate the drug exposure for individual dose adaptions would be of high clinical value. In this work, we propose a new technique using machine learning to estimate the tacrolimus exposure in kidney transplant recipients. Our models achieve predictive errors that are at the same level as an established population pharmacokinetic model, but are faster to develop and require less knowledge about the pharmacokinetic properties of the drug.
Assessing generalisability of deep learning-based polyp detection and segmentation methods through a computer vision challenge
Feb 24, 2022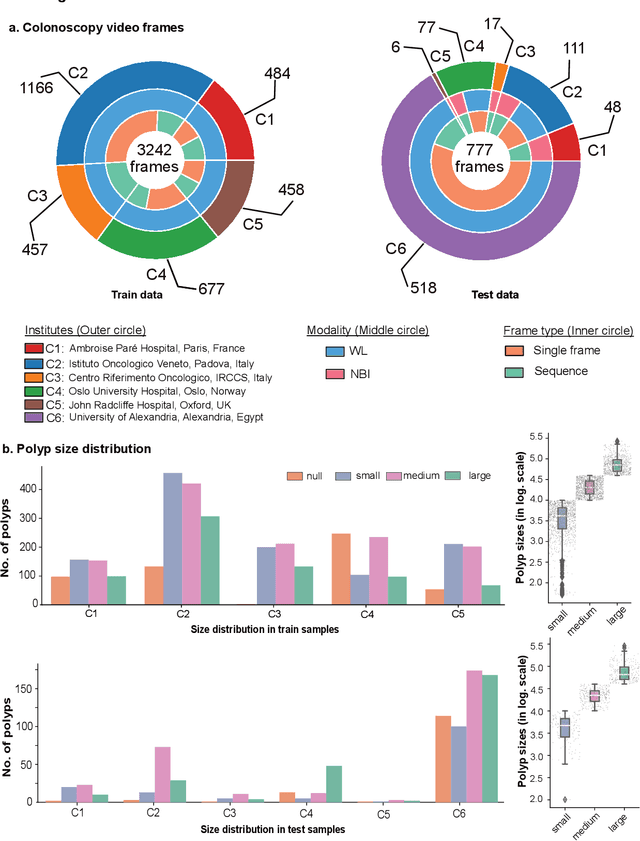
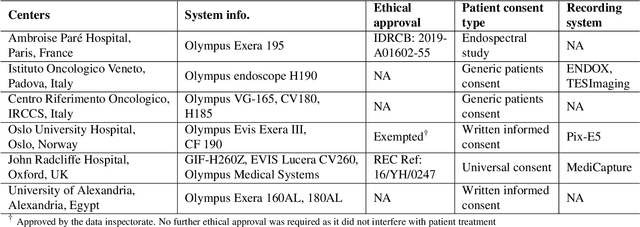
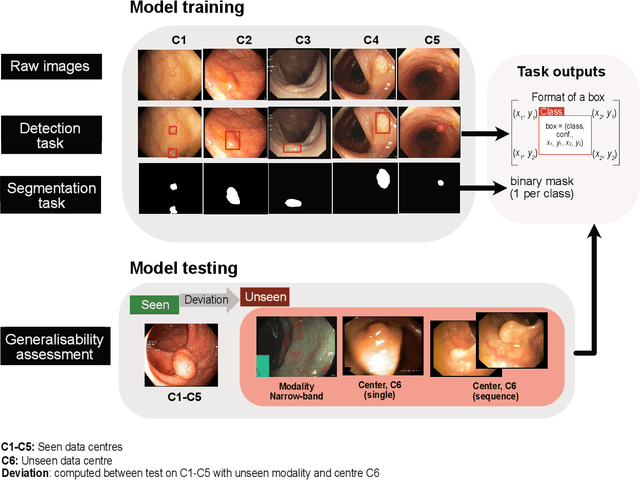
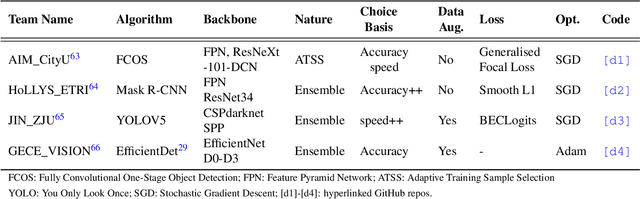
Abstract:Polyps are well-known cancer precursors identified by colonoscopy. However, variability in their size, location, and surface largely affect identification, localisation, and characterisation. Moreover, colonoscopic surveillance and removal of polyps (referred to as polypectomy ) are highly operator-dependent procedures. There exist a high missed detection rate and incomplete removal of colonic polyps due to their variable nature, the difficulties to delineate the abnormality, the high recurrence rates, and the anatomical topography of the colon. There have been several developments in realising automated methods for both detection and segmentation of these polyps using machine learning. However, the major drawback in most of these methods is their ability to generalise to out-of-sample unseen datasets that come from different centres, modalities and acquisition systems. To test this hypothesis rigorously we curated a multi-centre and multi-population dataset acquired from multiple colonoscopy systems and challenged teams comprising machine learning experts to develop robust automated detection and segmentation methods as part of our crowd-sourcing Endoscopic computer vision challenge (EndoCV) 2021. In this paper, we analyse the detection results of the four top (among seven) teams and the segmentation results of the five top teams (among 16). Our analyses demonstrate that the top-ranking teams concentrated on accuracy (i.e., accuracy > 80% on overall Dice score on different validation sets) over real-time performance required for clinical applicability. We further dissect the methods and provide an experiment-based hypothesis that reveals the need for improved generalisability to tackle diversity present in multi-centre datasets.
Visual Sentiment Analysis: A Natural DisasterUse-case Task at MediaEval 2021
Nov 22, 2021Abstract:The Visual Sentiment Analysis task is being offered for the first time at MediaEval. The main purpose of the task is to predict the emotional response to images of natural disasters shared on social media. Disaster-related images are generally complex and often evoke an emotional response, making them an ideal use case of visual sentiment analysis. We believe being able to perform meaningful analysis of natural disaster-related data could be of great societal importance, and a joint effort in this regard can open several interesting directions for future research. The task is composed of three sub-tasks, each aiming to explore a different aspect of the challenge. In this paper, we provide a detailed overview of the task, the general motivation of the task, and an overview of the dataset and the metrics to be used for the evaluation of the proposed solutions.
PAANet: Progressive Alternating Attention for Automatic Medical Image Segmentation
Nov 20, 2021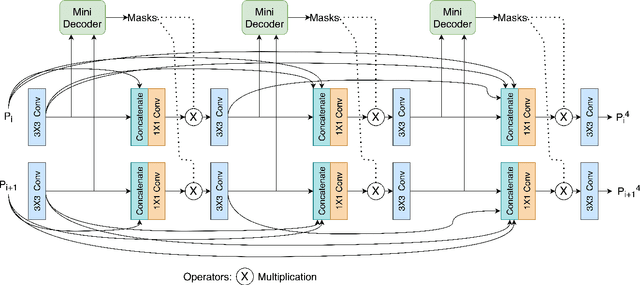
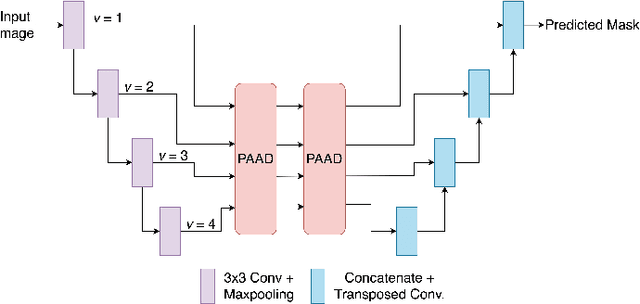
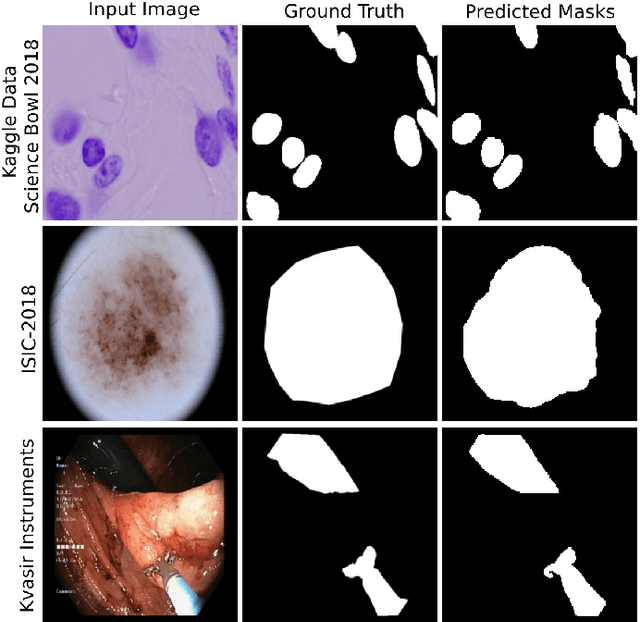
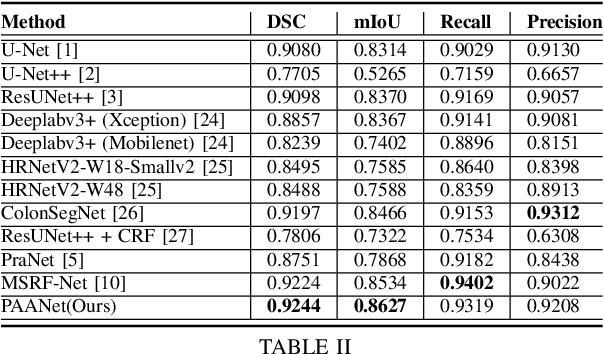
Abstract:Medical image segmentation can provide detailed information for clinical analysis which can be useful for scenarios where the detailed location of a finding is important. Knowing the location of disease can play a vital role in treatment and decision-making. Convolutional neural network (CNN) based encoder-decoder techniques have advanced the performance of automated medical image segmentation systems. Several such CNN-based methodologies utilize techniques such as spatial- and channel-wise attention to enhance performance. Another technique that has drawn attention in recent years is residual dense blocks (RDBs). The successive convolutional layers in densely connected blocks are capable of extracting diverse features with varied receptive fields and thus, enhancing performance. However, consecutive stacked convolutional operators may not necessarily generate features that facilitate the identification of the target structures. In this paper, we propose a progressive alternating attention network (PAANet). We develop progressive alternating attention dense (PAAD) blocks, which construct a guiding attention map (GAM) after every convolutional layer in the dense blocks using features from all scales. The GAM allows the following layers in the dense blocks to focus on the spatial locations relevant to the target region. Every alternate PAAD block inverts the GAM to generate a reverse attention map which guides ensuing layers to extract boundary and edge-related information, refining the segmentation process. Our experiments on three different biomedical image segmentation datasets exhibit that our PAANet achieves favourable performance when compared to other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge