Longjiang Zhang
Cardiac-CLIP: A Vision-Language Foundation Model for 3D Cardiac CT Images
Jul 29, 2025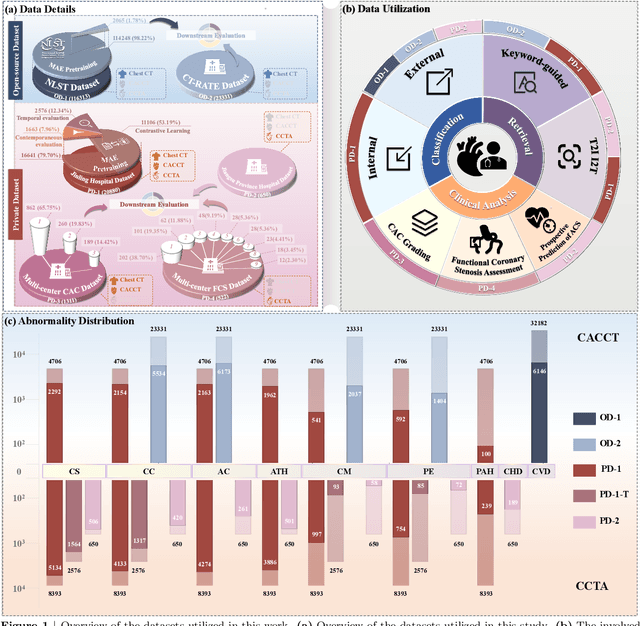
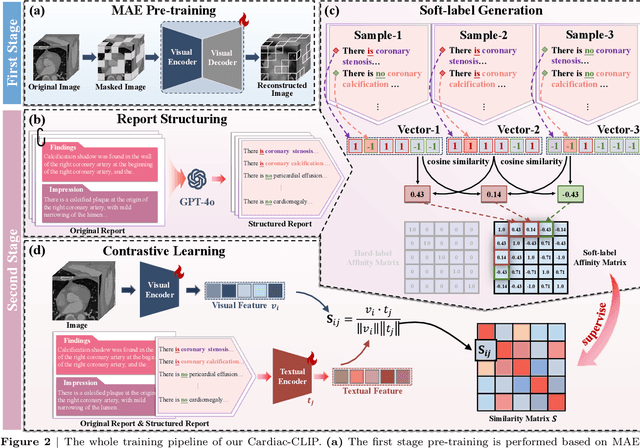
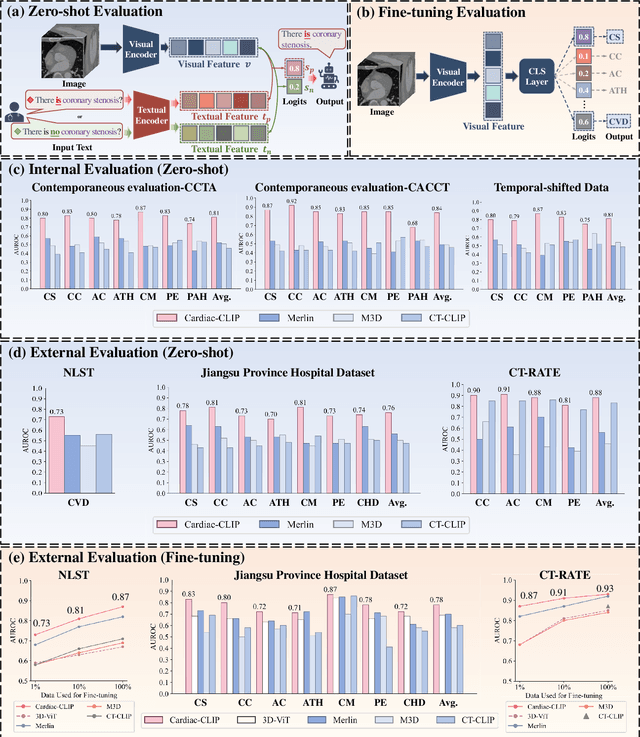
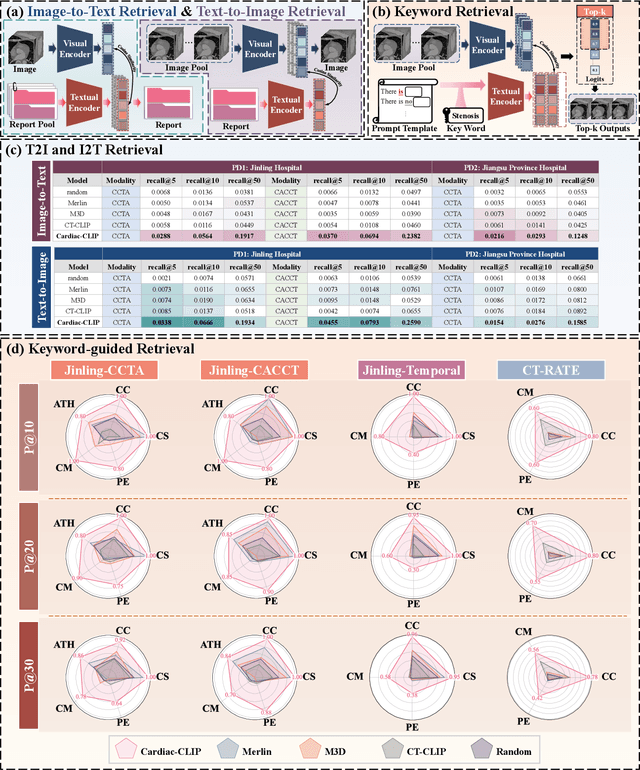
Abstract:Foundation models have demonstrated remarkable potential in medical domain. However, their application to complex cardiovascular diagnostics remains underexplored. In this paper, we present Cardiac-CLIP, a multi-modal foundation model designed for 3D cardiac CT images. Cardiac-CLIP is developed through a two-stage pre-training strategy. The first stage employs a 3D masked autoencoder (MAE) to perform self-supervised representation learning from large-scale unlabeled volumetric data, enabling the visual encoder to capture rich anatomical and contextual features. In the second stage, contrastive learning is introduced to align visual and textual representations, facilitating cross-modal understanding. To support the pre-training, we collect 16641 real clinical CT scans, supplemented by 114k publicly available data. Meanwhile, we standardize free-text radiology reports into unified templates and construct the pathology vectors according to diagnostic attributes, based on which the soft-label matrix is generated to supervise the contrastive learning process. On the other hand, to comprehensively evaluate the effectiveness of Cardiac-CLIP, we collect 6,722 real-clinical data from 12 independent institutions, along with the open-source data to construct the evaluation dataset. Specifically, Cardiac-CLIP is comprehensively evaluated across multiple tasks, including cardiovascular abnormality classification, information retrieval and clinical analysis. Experimental results demonstrate that Cardiac-CLIP achieves state-of-the-art performance across various downstream tasks in both internal and external data. Particularly, Cardiac-CLIP exhibits great effectiveness in supporting complex clinical tasks such as the prospective prediction of acute coronary syndrome, which is notoriously difficult in real-world scenarios.
FCaS: Fine-grained Cardiac Image Synthesis based on 3D Template Conditional Diffusion Model
Mar 12, 2025Abstract:Solving medical imaging data scarcity through semantic image generation has attracted significant attention in recent years. However, existing methods primarily focus on generating whole-organ or large-tissue structures, showing limited effectiveness for organs with fine-grained structure. Due to stringent topological consistency, fragile coronary features, and complex 3D morphological heterogeneity in cardiac imaging, accurately reconstructing fine-grained anatomical details of the heart remains a great challenge. To address this problem, in this paper, we propose the Fine-grained Cardiac image Synthesis(FCaS) framework, established on 3D template conditional diffusion model. FCaS achieves precise cardiac structure generation using Template-guided Conditional Diffusion Model (TCDM) through bidirectional mechanisms, which provides the fine-grained topological structure information of target image through the guidance of template. Meanwhile, we design a deformable Mask Generation Module (MGM) to mitigate the scarcity of high-quality and diverse reference mask in the generation process. Furthermore, to alleviate the confusion caused by imprecise synthetic images, we propose a Confidence-aware Adaptive Learning (CAL) strategy to facilitate the pre-training of downstream segmentation tasks. Specifically, we introduce the Skip-Sampling Variance (SSV) estimation to obtain confidence maps, which are subsequently employed to rectify the pre-training on downstream tasks. Experimental results demonstrate that images generated from FCaS achieves state-of-the-art performance in topological consistency and visual quality, which significantly facilitates the downstream tasks as well. Code will be released in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge