Juan C. Caicedo
Morgridge Institute for Research, Madison, WI, USA, University of Wisconsin-Madison, Madison, WI, USA
CHAMMI-75: pre-training multi-channel models with heterogeneous microscopy images
Dec 23, 2025Abstract:Quantifying cell morphology using images and machine learning has proven to be a powerful tool to study the response of cells to treatments. However, models used to quantify cellular morphology are typically trained with a single microscopy imaging type. This results in specialized models that cannot be reused across biological studies because the technical specifications do not match (e.g., different number of channels), or because the target experimental conditions are out of distribution. Here, we present CHAMMI-75, an open access dataset of heterogeneous, multi-channel microscopy images from 75 diverse biological studies. We curated this resource from publicly available sources to investigate cellular morphology models that are channel-adaptive and can process any microscopy image type. Our experiments show that training with CHAMMI-75 can improve performance in multi-channel bioimaging tasks primarily because of its high diversity in microscopy modalities. This work paves the way to create the next generation of cellular morphology models for biological studies.
ChA-MAEViT: Unifying Channel-Aware Masked Autoencoders and Multi-Channel Vision Transformers for Improved Cross-Channel Learning
Mar 25, 2025Abstract:Prior work using Masked Autoencoders (MAEs) typically relies on random patch masking based on the assumption that images have significant redundancies across different channels, allowing for the reconstruction of masked content using cross-channel correlations. However, this assumption does not hold in Multi-Channel Imaging (MCI), where channels may provide complementary information with minimal feature overlap. Thus, these MAEs primarily learn local structures within individual channels from patch reconstruction, failing to fully leverage cross-channel interactions and limiting their MCI effectiveness. In this paper, we present ChA-MAEViT, an MAE-based method that enhances feature learning across MCI channels via four key strategies: (1) dynamic channel-patch masking, which compels the model to reconstruct missing channels in addition to masked patches, thereby enhancing cross-channel dependencies and improving robustness to varying channel configurations; (2) memory tokens, which serve as long-term memory aids to promote information sharing across channels, addressing the challenges of reconstructing structurally diverse channels; (3) hybrid token fusion module, which merges fine-grained patch tokens with a global class token to capture richer representations; and (4) Channel-Aware Decoder, a lightweight decoder utilizes channel tokens to effectively reconstruct image patches. Experiments on satellite and microscopy datasets, CHAMMI, JUMP-CP, and So2Sat, show that ChA-MAEViT significantly outperforms state-of-the-art MCI-ViTs by 3.0-21.5%, highlighting the importance of cross-channel interactions in MCI.
A Time-Intensity Aware Pipeline for Generating Late-Stage Breast DCE-MRI using Generative Adversarial Models
Sep 03, 2024Abstract:Contrast-enhancement pattern analysis is critical in breast magnetic resonance imaging (MRI) to distinguish benign from probably malignant tumors. However, contrast-enhanced image acquisitions are time-consuming and very expensive. As an alternative to physical acquisition, this paper proposes a comprehensive pipeline for the generation of accurate long-term (late) contrast-enhanced breast MRI from the early counterpart. The proposed strategy focuses on preserving the contrast agent pattern in the enhanced regions while maintaining visual properties in the entire synthesized images. To that end, a novel loss function that leverages the biological behavior of contrast agent (CA) in tissue, given by the Time-Intensity (TI) enhancement curve, is proposed to optimize a pixel-attention based generative model. In addition, unlike traditional normalization and standardization methods, we developed a new normalization strategy that maintains the contrast enhancement pattern across the image sequences at multiple timestamps. This ensures the prevalence of the CA pattern after image preprocessing, unlike conventional approaches. Furthermore, in order to objectively evaluate the clinical quality of the synthesized images, two metrics are also introduced to measure the differences between the TI curves of enhanced regions of the acquired and synthesized images. The experimental results showed that the proposed strategy generates images that significantly outperform diagnostic quality in contrast-enhanced regions while maintaining the spatial features of the entire image. This results suggest a potential use of synthetic late enhanced images generated via deep learning in clinical scenarios.
CHAMMI: A benchmark for channel-adaptive models in microscopy imaging
Oct 30, 2023



Abstract:Most neural networks assume that input images have a fixed number of channels (three for RGB images). However, there are many settings where the number of channels may vary, such as microscopy images where the number of channels changes depending on instruments and experimental goals. Yet, there has not been a systemic attempt to create and evaluate neural networks that are invariant to the number and type of channels. As a result, trained models remain specific to individual studies and are hardly reusable for other microscopy settings. In this paper, we present a benchmark for investigating channel-adaptive models in microscopy imaging, which consists of 1) a dataset of varied-channel single-cell images, and 2) a biologically relevant evaluation framework. In addition, we adapted several existing techniques to create channel-adaptive models and compared their performance on this benchmark to fixed-channel, baseline models. We find that channel-adaptive models can generalize better to out-of-domain tasks and can be computationally efficient. We contribute a curated dataset (https://doi.org/10.5281/zenodo.7988357) and an evaluation API (https://github.com/broadinstitute/MorphEm.git) to facilitate objective comparisons in future research and applications.
Out of Distribution Generalization via Interventional Style Transfer in Single-Cell Microscopy
Jun 15, 2023



Abstract:Real-world deployment of computer vision systems, including in the discovery processes of biomedical research, requires causal representations that are invariant to contextual nuisances and generalize to new data. Leveraging the internal replicate structure of two novel single-cell fluorescent microscopy datasets, we propose generally applicable tests to assess the extent to which models learn causal representations across increasingly challenging levels of OOD-generalization. We show that despite seemingly strong performance, as assessed by other established metrics, both naive and contemporary baselines designed to ward against confounding, collapse on these tests. We introduce a new method, Interventional Style Transfer (IST), that substantially improves OOD generalization by generating interventional training distributions in which spurious correlations between biological causes and nuisances are mitigated. We publish our code and datasets.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Anchoring to Exemplars for Training Mixture-of-Expert Cell Embeddings
Dec 06, 2021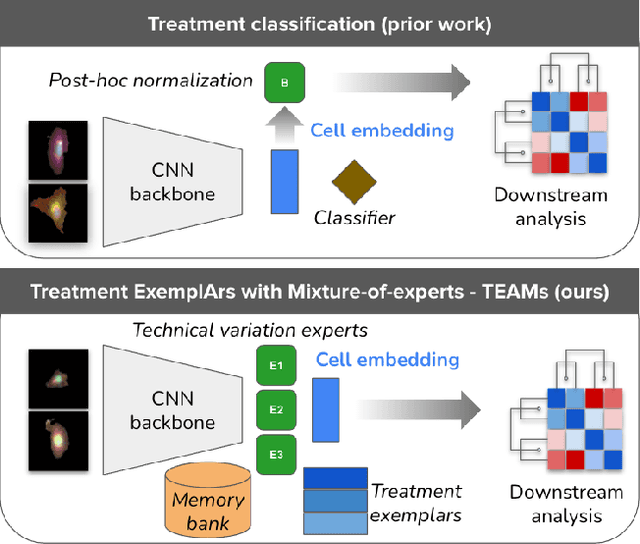
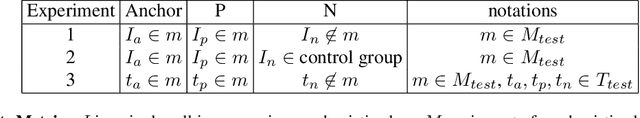
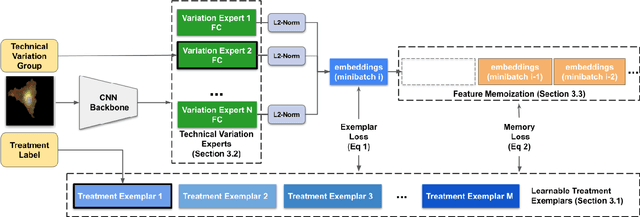
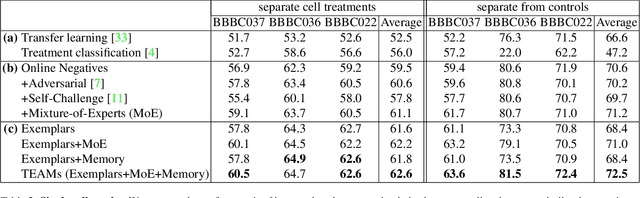
Abstract:Analyzing the morphology of cells in microscopy images can provide insights into the mechanism of compounds or the function of genes. Addressing this task requires methods that can not only extract biological information from the images, but also ignore technical variations, ie, changes in experimental procedure or differences between equipments used to collect microscopy images. We propose Treatment ExemplArs with Mixture-of-experts (TEAMs), an embedding learning approach that learns a set of experts that are specialized in capturing technical variations in our training set and then aggregates specialist's predictions at test time. Thus, TEAMs can learn powerful embeddings with less technical variation bias by minimizing the noise from every expert. To train our model, we leverage Treatment Exemplars that enable our approach to capture the distribution of the entire dataset in every minibatch while still fitting into GPU memory. We evaluate our approach on three datasets for tasks like drug discovery, boosting performance on identifying the true mechanism of action of cell treatments by 5.5-11% over the state-of-the-art.
Quantum Latent Semantic Analysis
Mar 07, 2019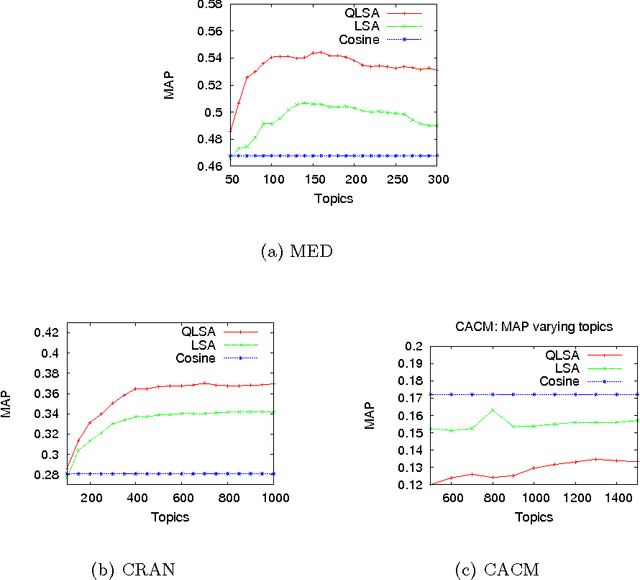
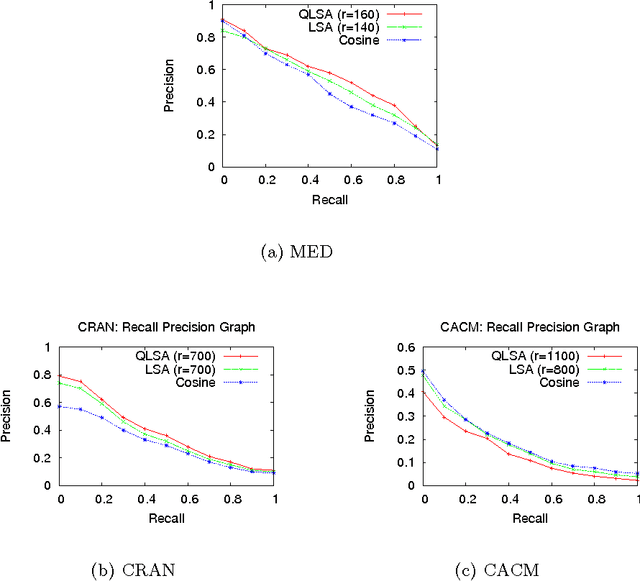
Abstract:The main goal of this paper is to explore latent topic analysis (LTA), in the context of quantum information retrieval. LTA is a valuable technique for document analysis and representation, which has been extensively used in information retrieval and machine learning. Different LTA techniques have been proposed, some based on geometrical modeling (such as latent semantic analysis, LSA) and others based on a strong statistical foundation. However, these two different approaches are not usually mixed. Quantum information retrieval has the remarkable virtue of combining both geometry and probability in a common principled framework. We built on this quantum framework to propose a new LTA method, which has a clear geometrical motivation but also supports a well-founded probabilistic interpretation. An initial exploratory experimentation was performed on three standard data sets. The results show that the proposed method outperforms LSA on two of the three datasets. These results suggests that the quantum-motivated representation is an alternative for geometrical latent topic modeling worthy of further exploration.
Flickr30k Entities: Collecting Region-to-Phrase Correspondences for Richer Image-to-Sentence Models
Sep 19, 2016
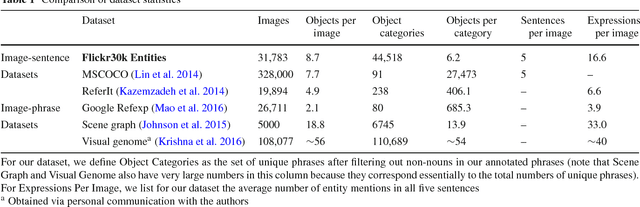
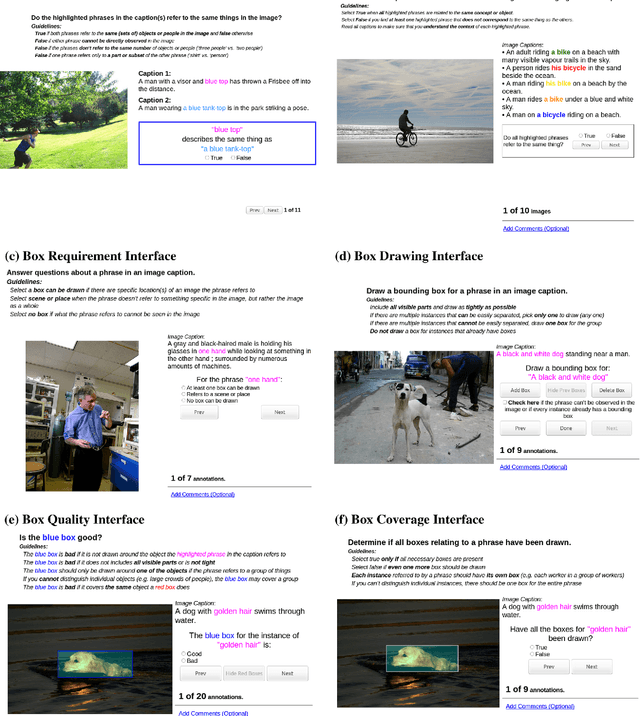

Abstract:The Flickr30k dataset has become a standard benchmark for sentence-based image description. This paper presents Flickr30k Entities, which augments the 158k captions from Flickr30k with 244k coreference chains, linking mentions of the same entities across different captions for the same image, and associating them with 276k manually annotated bounding boxes. Such annotations are essential for continued progress in automatic image description and grounded language understanding. They enable us to define a new benchmark for localization of textual entity mentions in an image. We present a strong baseline for this task that combines an image-text embedding, detectors for common objects, a color classifier, and a bias towards selecting larger objects. While our baseline rivals in accuracy more complex state-of-the-art models, we show that its gains cannot be easily parlayed into improvements on such tasks as image-sentence retrieval, thus underlining the limitations of current methods and the need for further research.
Active Object Localization with Deep Reinforcement Learning
Nov 18, 2015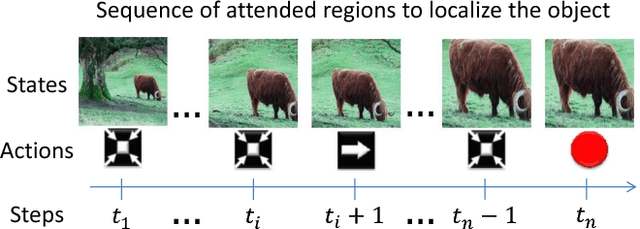
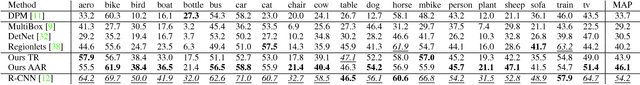

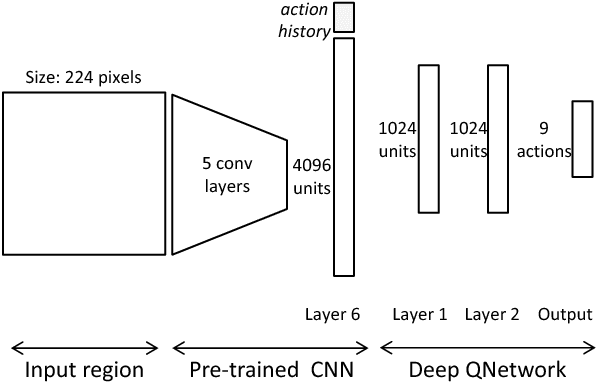
Abstract:We present an active detection model for localizing objects in scenes. The model is class-specific and allows an agent to focus attention on candidate regions for identifying the correct location of a target object. This agent learns to deform a bounding box using simple transformation actions, with the goal of determining the most specific location of target objects following top-down reasoning. The proposed localization agent is trained using deep reinforcement learning, and evaluated on the Pascal VOC 2007 dataset. We show that agents guided by the proposed model are able to localize a single instance of an object after analyzing only between 11 and 25 regions in an image, and obtain the best detection results among systems that do not use object proposals for object localization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge