Nikita Moshkov
Institute of Computational Biology, Helmholtz Munich, Neuherberg, Germany
CHAMMI-75: pre-training multi-channel models with heterogeneous microscopy images
Dec 23, 2025Abstract:Quantifying cell morphology using images and machine learning has proven to be a powerful tool to study the response of cells to treatments. However, models used to quantify cellular morphology are typically trained with a single microscopy imaging type. This results in specialized models that cannot be reused across biological studies because the technical specifications do not match (e.g., different number of channels), or because the target experimental conditions are out of distribution. Here, we present CHAMMI-75, an open access dataset of heterogeneous, multi-channel microscopy images from 75 diverse biological studies. We curated this resource from publicly available sources to investigate cellular morphology models that are channel-adaptive and can process any microscopy image type. Our experiments show that training with CHAMMI-75 can improve performance in multi-channel bioimaging tasks primarily because of its high diversity in microscopy modalities. This work paves the way to create the next generation of cellular morphology models for biological studies.
CHAMMI: A benchmark for channel-adaptive models in microscopy imaging
Oct 30, 2023



Abstract:Most neural networks assume that input images have a fixed number of channels (three for RGB images). However, there are many settings where the number of channels may vary, such as microscopy images where the number of channels changes depending on instruments and experimental goals. Yet, there has not been a systemic attempt to create and evaluate neural networks that are invariant to the number and type of channels. As a result, trained models remain specific to individual studies and are hardly reusable for other microscopy settings. In this paper, we present a benchmark for investigating channel-adaptive models in microscopy imaging, which consists of 1) a dataset of varied-channel single-cell images, and 2) a biologically relevant evaluation framework. In addition, we adapted several existing techniques to create channel-adaptive models and compared their performance on this benchmark to fixed-channel, baseline models. We find that channel-adaptive models can generalize better to out-of-domain tasks and can be computationally efficient. We contribute a curated dataset (https://doi.org/10.5281/zenodo.7988357) and an evaluation API (https://github.com/broadinstitute/MorphEm.git) to facilitate objective comparisons in future research and applications.
Anchoring to Exemplars for Training Mixture-of-Expert Cell Embeddings
Dec 06, 2021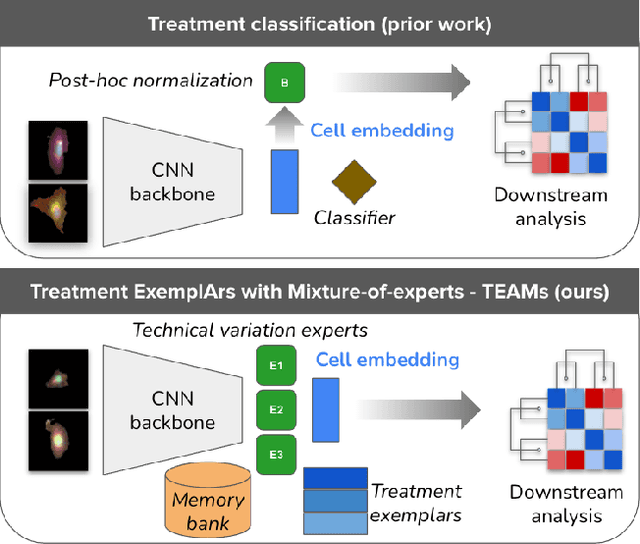
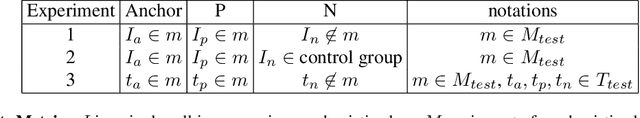
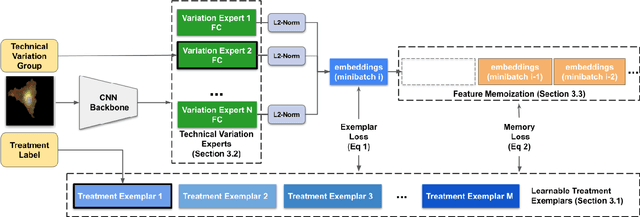
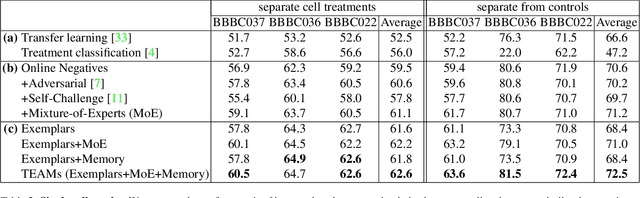
Abstract:Analyzing the morphology of cells in microscopy images can provide insights into the mechanism of compounds or the function of genes. Addressing this task requires methods that can not only extract biological information from the images, but also ignore technical variations, ie, changes in experimental procedure or differences between equipments used to collect microscopy images. We propose Treatment ExemplArs with Mixture-of-experts (TEAMs), an embedding learning approach that learns a set of experts that are specialized in capturing technical variations in our training set and then aggregates specialist's predictions at test time. Thus, TEAMs can learn powerful embeddings with less technical variation bias by minimizing the noise from every expert. To train our model, we leverage Treatment Exemplars that enable our approach to capture the distribution of the entire dataset in every minibatch while still fitting into GPU memory. We evaluate our approach on three datasets for tasks like drug discovery, boosting performance on identifying the true mechanism of action of cell treatments by 5.5-11% over the state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge