Jeremy Tan
Accelerating Antimicrobial Discovery with Controllable Deep Generative Models and Molecular Dynamics
May 22, 2020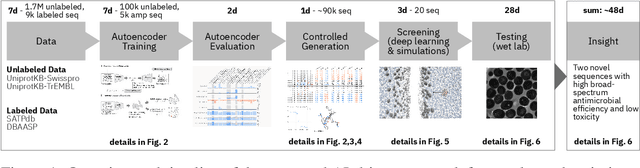
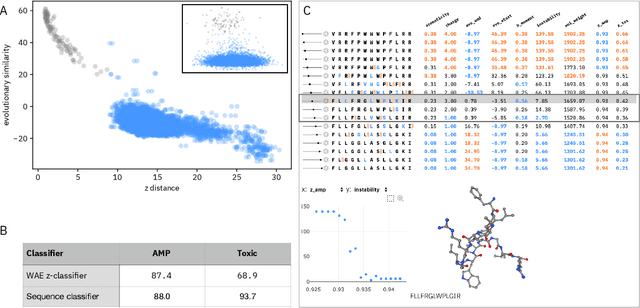
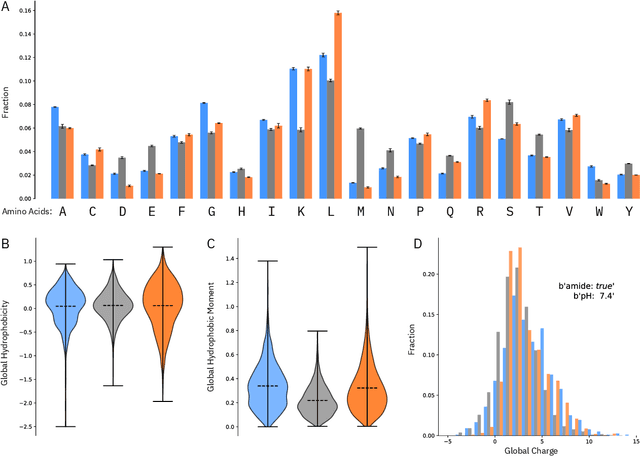
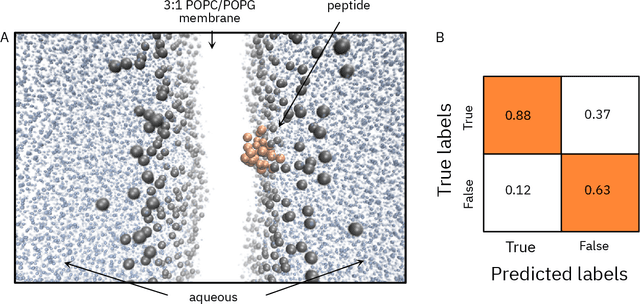
Abstract:De novo therapeutic design is challenged by a vast chemical repertoire and multiple constraints such as high broad-spectrum potency and low toxicity. We propose CLaSS (Controlled Latent attribute Space Sampling) - a novel and efficient computational method for attribute-controlled generation of molecules, which leverages guidance from classifiers trained on an informative latent space of molecules modeled using a deep generative autoencoder. We further screen the generated molecules by using a set of deep learning classifiers in conjunction with novel physicochemical features derived from high-throughput molecular simulations. The proposed approach is employed for designing non-toxic antimicrobial peptides (AMPs) with strong broad-spectrum potency, which are emerging drug candidates for tackling antibiotic resistance. Synthesis and wet lab testing of only twenty designed sequences identified two novel and minimalist AMPs with high potency against diverse Gram-positive and Gram-negative pathogens, including the hard-to-treat multidrug-resistant K. pneumoniae, as well as low in vitro and in vivo toxicity. The proposed approach thus presents a viable path for faster discovery of potent and selective broad-spectrum antimicrobials with a higher success rate than state-of-the-art methods.
Ultrasound Video Summarization using Deep Reinforcement Learning
May 19, 2020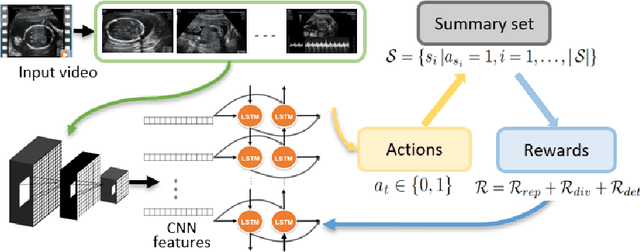
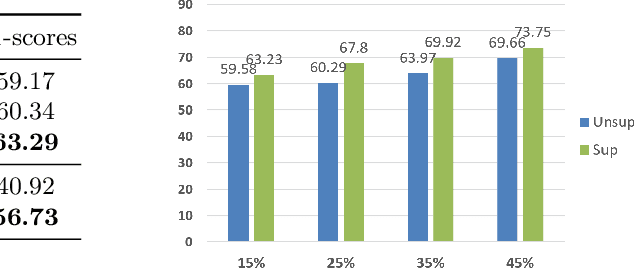
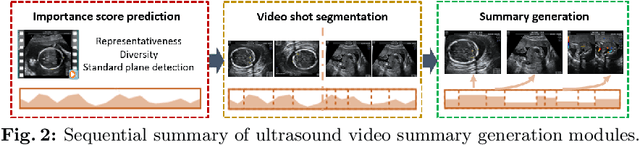
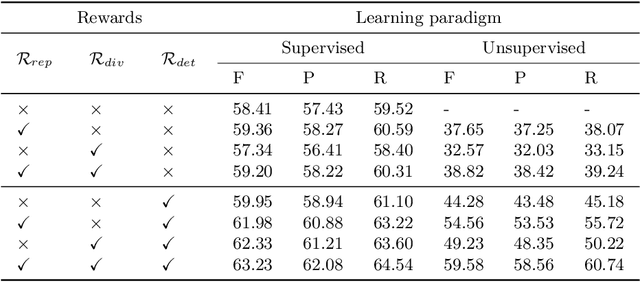
Abstract:Video is an essential imaging modality for diagnostics, e.g. in ultrasound imaging, for endoscopy, or movement assessment. However, video hasn't received a lot of attention in the medical image analysis community. In the clinical practice, it is challenging to utilise raw diagnostic video data efficiently as video data takes a long time to process, annotate or audit. In this paper we introduce a novel, fully automatic video summarization method that is tailored to the needs of medical video data. Our approach is framed as reinforcement learning problem and produces agents focusing on the preservation of important diagnostic information. We evaluate our method on videos from fetal ultrasound screening, where commonly only a small amount of the recorded data is used diagnostically. We show that our method is superior to alternative video summarization methods and that it preserves essential information required by clinical diagnostic standards.
Divergent Search for Few-Shot Image Classification
Apr 16, 2020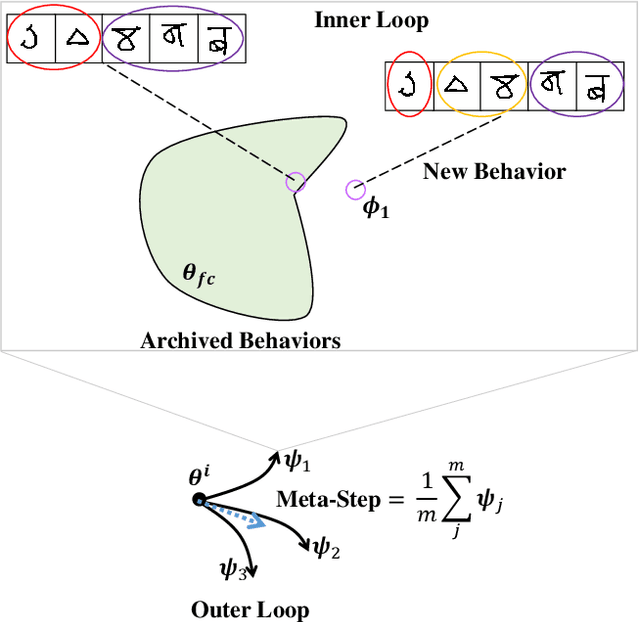
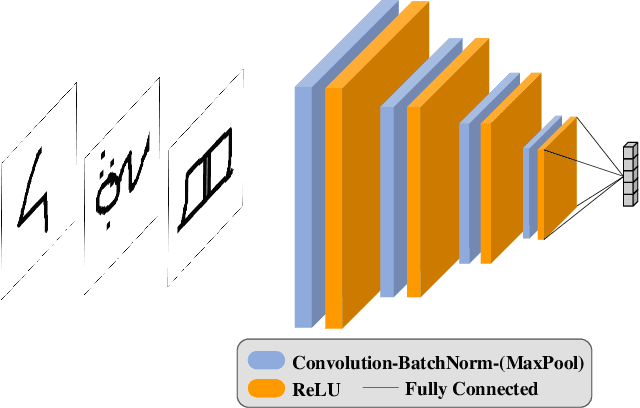
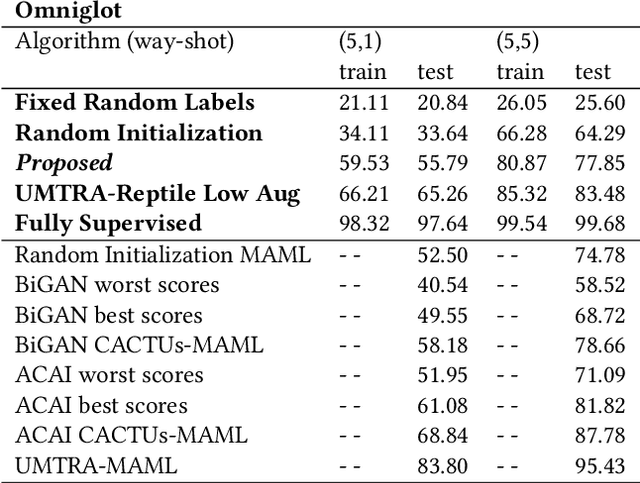
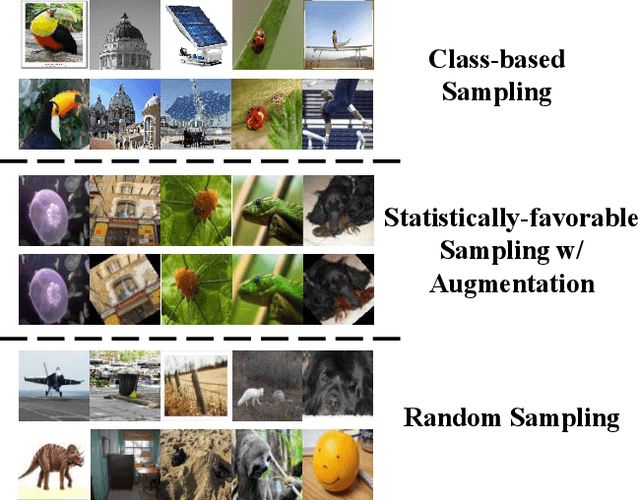
Abstract:When data is unlabelled and the target task is not known a priori, divergent search offers a strategy for learning a wide range of skills. Having such a repertoire allows a system to adapt to new, unforeseen tasks. Unlabelled image data is plentiful, but it is not always known which features will be required for downstream tasks. We propose a method for divergent search in the few-shot image classification setting and evaluate with Omniglot and Mini-ImageNet. This high-dimensional behavior space includes all possible ways of partitioning the data. To manage divergent search in this space, we rely on a meta-learning framework to integrate useful features from diverse tasks into a single model. The final layer of this model is used as an index into the `archive' of all past behaviors. We search for regions in the behavior space that the current archive cannot reach. As expected, divergent search is outperformed by models with a strong bias toward the evaluation tasks. But it is able to match and sometimes exceed the performance of models that have a weak bias toward the target task or none at all. This demonstrates that divergent search is a viable approach, even in high-dimensional behavior spaces.
Semi-supervised Learning of Fetal Anatomy from Ultrasound
Aug 30, 2019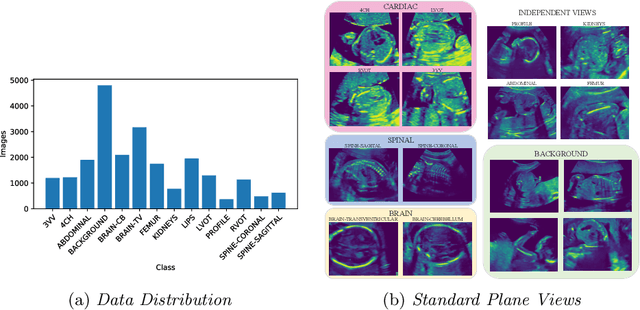
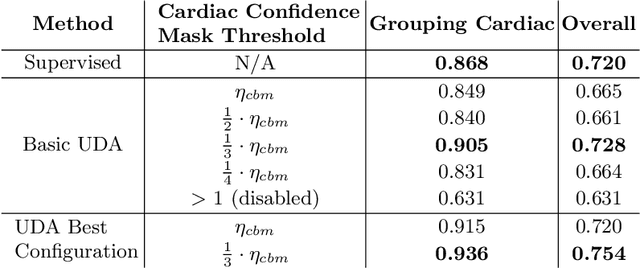
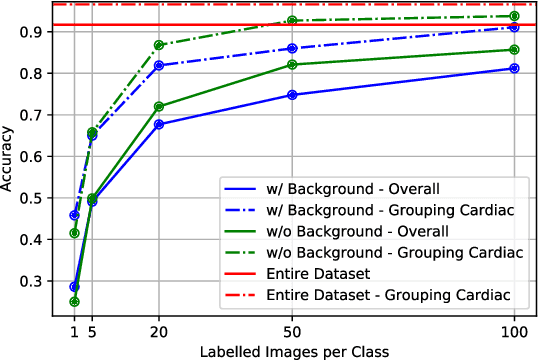
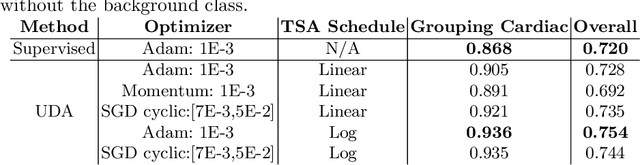
Abstract:Semi-supervised learning methods have achieved excellent performance on standard benchmark datasets using very few labelled images. Anatomy classification in fetal 2D ultrasound is an ideal problem setting to test whether these results translate to non-ideal data. Our results indicate that inclusion of a challenging background class can be detrimental and that semi-supervised learning mostly benefits classes that are already distinct, sometimes at the expense of more similar classes.
Morpho-MNIST: Quantitative Assessment and Diagnostics for Representation Learning
Sep 27, 2018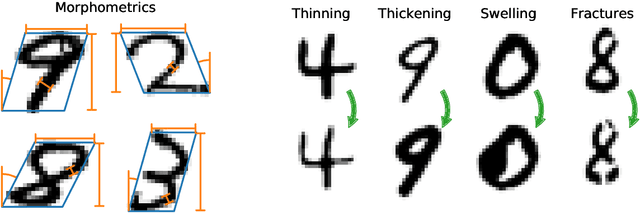

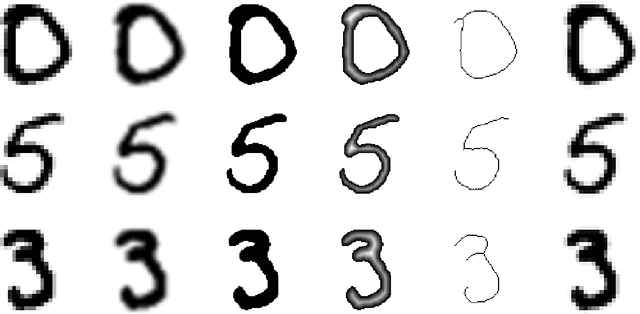

Abstract:Revealing latent structure in data is an active field of research, having brought exciting new models such as variational autoencoders and generative adversarial networks, and is essential to push machine learning towards unsupervised knowledge discovery. However, a major challenge is the lack of suitable benchmarks for an objective and quantitative evaluation of learned representations. To address this issue we introduce Morpho-MNIST. We extend the popular MNIST dataset by adding a morphometric analysis enabling quantitative comparison of different models, identification of the roles of latent variables, and characterisation of sample diversity. We further propose a set of quantifiable perturbations to assess the performance of unsupervised and supervised methods on challenging tasks such as outlier detection and domain adaptation.
Deep Adversarial Context-Aware Landmark Detection for Ultrasound Imaging
May 28, 2018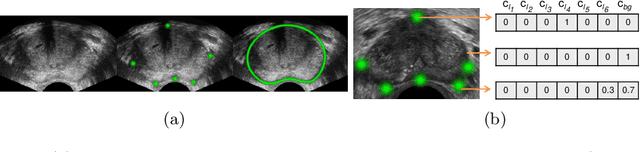
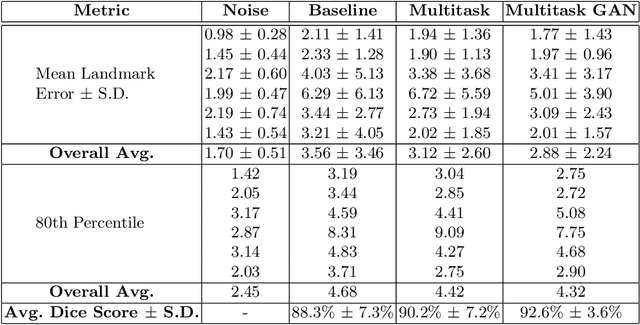
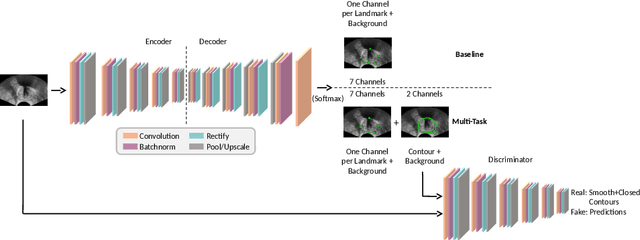
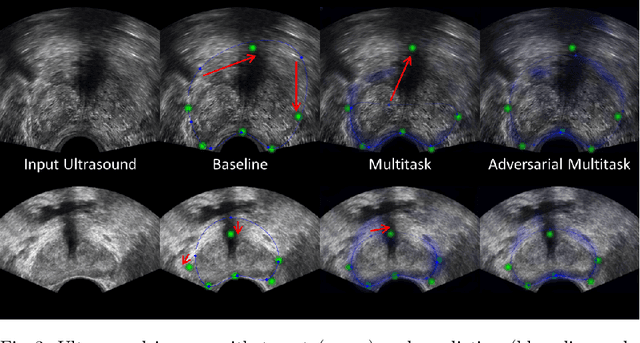
Abstract:Real-time localization of prostate gland in trans-rectal ultrasound images is a key technology that is required to automate the ultrasound guided prostate biopsy procedures. In this paper, we propose a new deep learning based approach which is aimed at localizing several prostate landmarks efficiently and robustly. We propose a multitask learning approach primarily to make the overall algorithm more contextually aware. In this approach, we not only consider the explicit learning of landmark locations, but also build-in a mechanism to learn the contour of the prostate. This multitask learning is further coupled with an adversarial arm to promote the generation of feasible structures. We have trained this network using ~4000 labeled trans-rectal ultrasound images and tested on an independent set of images with ground truth landmark locations. We have achieved an overall Dice score of 92.6% for the adversarially trained multitask approach, which is significantly better than the Dice score of 88.3% obtained by only learning of landmark locations. The overall mean distance error using the adversarial multitask approach has also improved by 20% while reducing the standard deviation of the error compared to learning landmark locations only. In terms of computational complexity both approaches can process the images in real-time using standard computer with a standard CUDA enabled GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge