Jens Rittscher
FANet: A Feedback Attention Network for Improved Biomedical Image Segmentation
Mar 31, 2021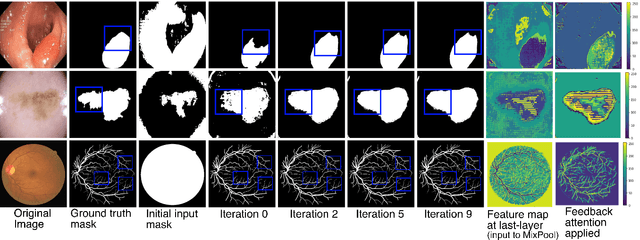
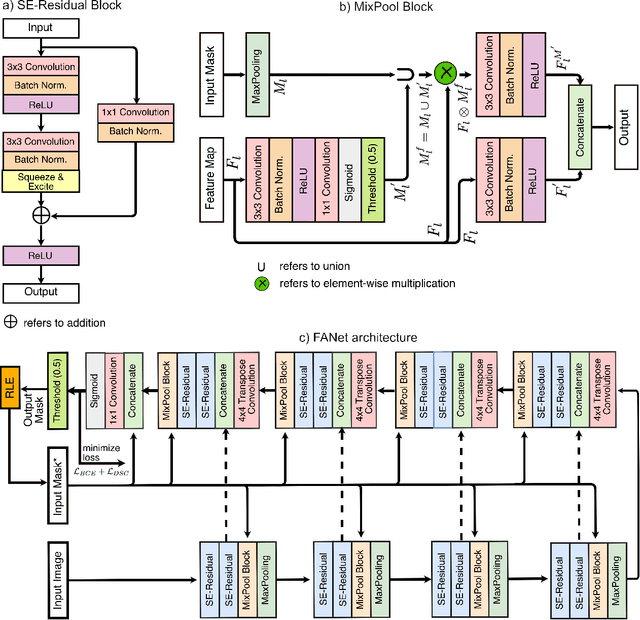
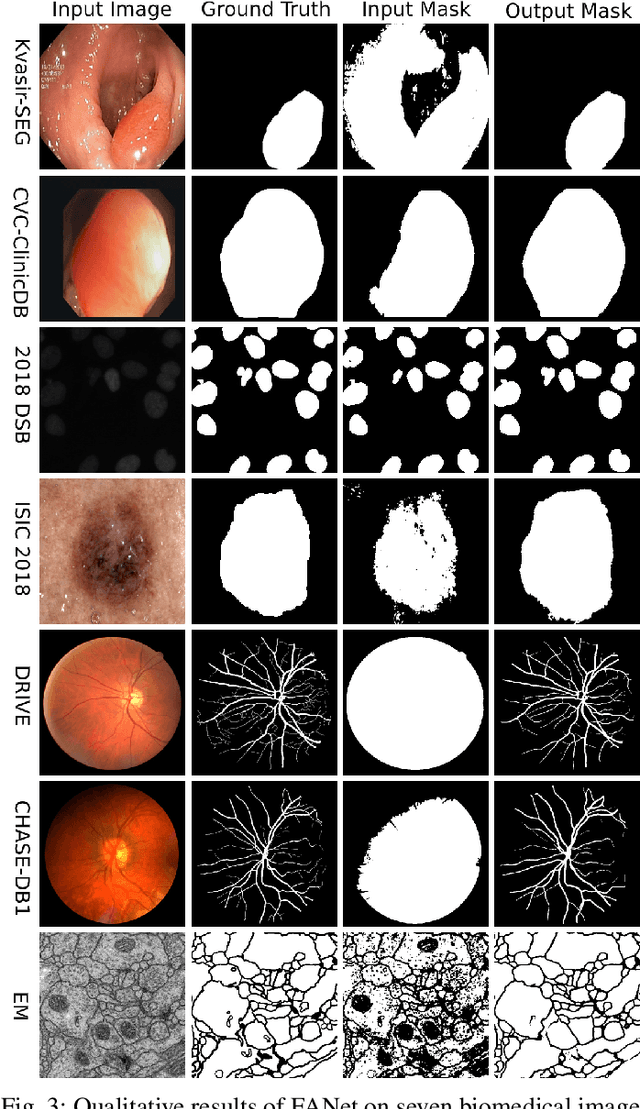
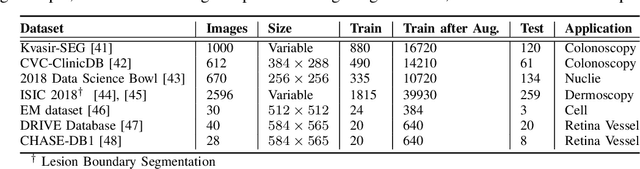
Abstract:With the increase in available large clinical and experimental datasets, there has been substantial amount of work being done on addressing the challenges in the area of biomedical image analysis. Image segmentation, which is crucial for any quantitative analysis, has especially attracted attention. Recent hardware advancement has led to the success of deep learning approaches. However, although deep learning models are being trained on large datasets, existing methods do not use the information from different learning epochs effectively. In this work, we leverage the information of each training epoch to prune the prediction maps of the subsequent epochs. We propose a novel architecture called feedback attention network (FANet) that unifies the previous epoch mask with the feature map of the current training epoch. The previous epoch mask is then used to provide a hard attention to the learnt feature maps at different convolutional layers. The network also allows to rectify the predictions in an iterative fashion during the test time. We show that our proposed feedback attention model provides a substantial improvement on most segmentation metrics tested on seven publicly available biomedical imaging datasets demonstrating the effectiveness of the proposed FANet.
Unsupervised Adversarial Domain Adaptation For Barrett's Segmentation
Dec 09, 2020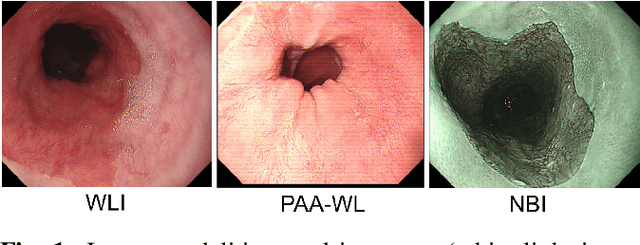
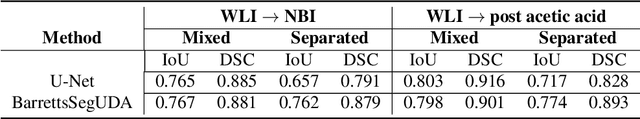
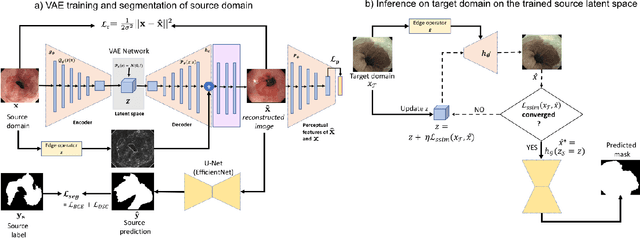
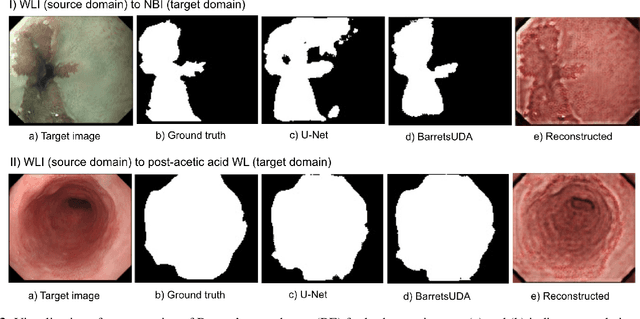
Abstract:Barrett's oesophagus (BE) is one of the early indicators of esophageal cancer. Patients with BE are monitored and undergo ablation therapies to minimise the risk, thereby making it eminent to identify the BE area precisely. Automated segmentation can help clinical endoscopists to assess and treat BE area more accurately. Endoscopy imaging of BE can include multiple modalities in addition to the conventional white light (WL) modality. Supervised models require large amount of manual annotations incorporating all data variability in the training data. However, it becomes cumbersome, tedious and labour intensive work to generate manual annotations, and additionally modality specific expertise is required. In this work, we aim to alleviate this problem by applying an unsupervised domain adaptation technique (UDA). Here, UDA is trained on white light endoscopy images as source domain and are well-adapted to generalise to produce segmentation on different imaging modalities as target domain, namely narrow band imaging and post acetic-acid WL imaging. Our dataset consists of a total of 871 images consisting of both source and target domains. Our results show that the UDA-based approach outperforms traditional supervised U-Net segmentation by nearly 10% on both Dice similarity coefficient and intersection-over-union.
Real-Time Polyp Detection, Localisation and Segmentation in Colonoscopy Using Deep Learning
Nov 15, 2020
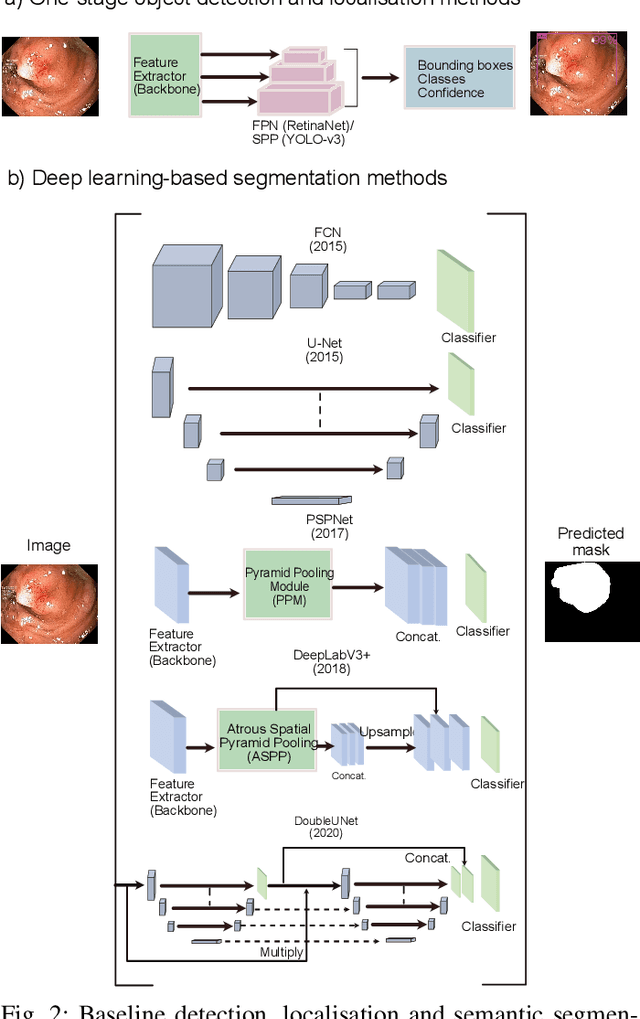
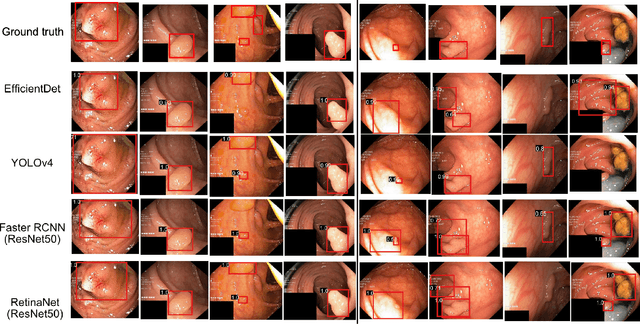

Abstract:Computer-aided detection, localisation, and segmentation methods can help improve colonoscopy procedures. Even though many methods have been built to tackle automatic detection and segmentation of polyps, benchmarking of state-of-the-art methods still remains an open problem. This is due to the increasing number of researched computer-vision methods that can be applied to polyp datasets. Benchmarking of novel methods can provide a direction to the development of automated polyp detection and segmentation tasks. Furthermore, it ensures that the produced results in the community are reproducible and provide a fair comparison of developed methods. In this paper, we benchmark several recent state-of-the-art methods using Kvasir-SEG, an open-access dataset of colonoscopy images, for polyp detection, localisation, and segmentation evaluating both method accuracy and speed. Whilst, most methods in literature have competitive performance over accuracy, we show that YOLOv4 with a Darknet53 backbone and cross-stage-partial connections achieved a better trade-off between an average precision of 0.8513 and mean IoU of 0.8025, and the fastest speed of 48 frames per second for the detection and localisation task. Likewise, UNet with a ResNet34 backbone achieved the highest dice coefficient of 0.8757 and the best average speed of 35 frames per second for the segmentation task. Our comprehensive comparison with various state-of-the-art methods reveal the importance of benchmarking the deep learning methods for automated real-time polyp identification and delineations that can potentially transform current clinical practices and minimise miss-detection rates.
A translational pathway of deep learning methods in GastroIntestinal Endoscopy
Oct 12, 2020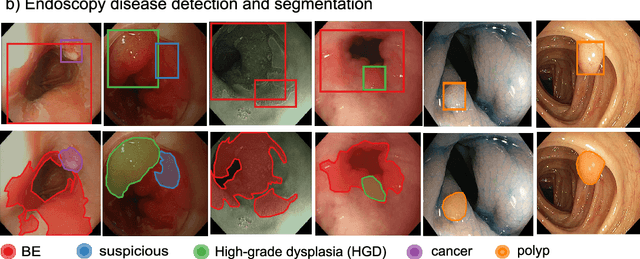
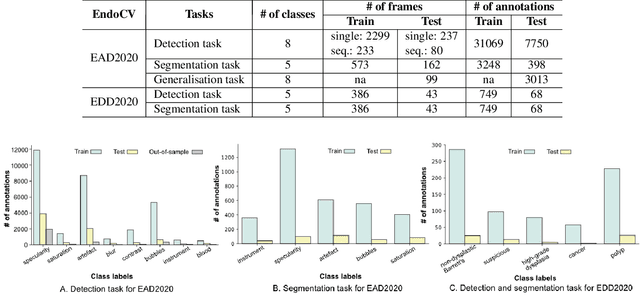
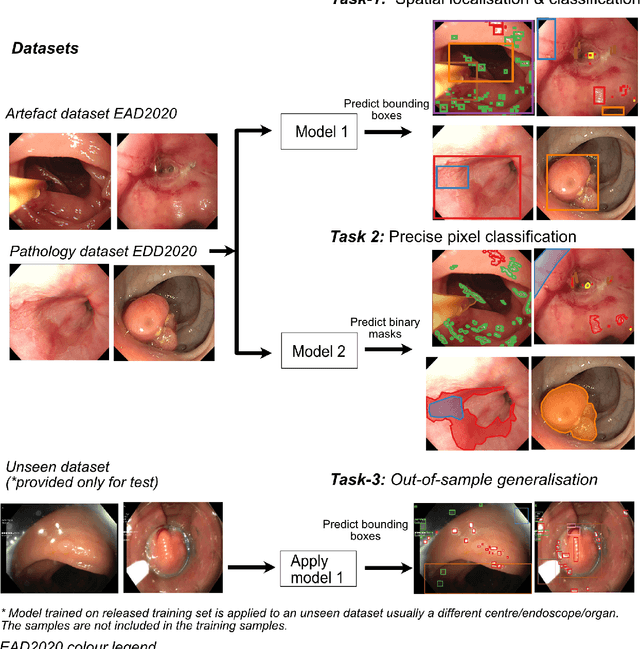
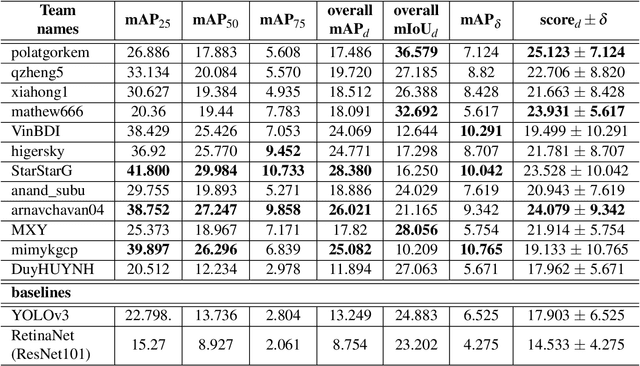
Abstract:The Endoscopy Computer Vision Challenge (EndoCV) is a crowd-sourcing initiative to address eminent problems in developing reliable computer aided detection and diagnosis endoscopy systems and suggest a pathway for clinical translation of technologies. Whilst endoscopy is a widely used diagnostic and treatment tool for hollow-organs, there are several core challenges often faced by endoscopists, mainly: 1) presence of multi-class artefacts that hinder their visual interpretation, and 2) difficulty in identifying subtle precancerous precursors and cancer abnormalities. Artefacts often affect the robustness of deep learning methods applied to the gastrointestinal tract organs as they can be confused with tissue of interest. EndoCV2020 challenges are designed to address research questions in these remits. In this paper, we present a summary of methods developed by the top 17 teams and provide an objective comparison of state-of-the-art methods and methods designed by the participants for two sub-challenges: i) artefact detection and segmentation (EAD2020), and ii) disease detection and segmentation (EDD2020). Multi-center, multi-organ, multi-class, and multi-modal clinical endoscopy datasets were compiled for both EAD2020 and EDD2020 sub-challenges. An out-of-sample generalisation ability of detection algorithms was also evaluated. Whilst most teams focused on accuracy improvements, only a few methods hold credibility for clinical usability. The best performing teams provided solutions to tackle class imbalance, and variabilities in size, origin, modality and occurrences by exploring data augmentation, data fusion, and optimal class thresholding techniques.
Microscopic fine-grained instance classification through deep attention
Oct 06, 2020
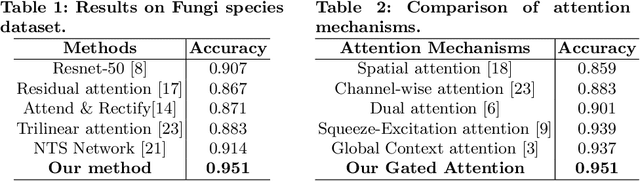
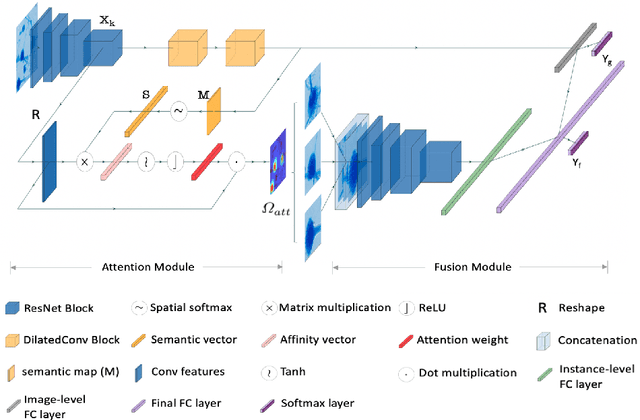
Abstract:Fine-grained classification of microscopic image data with limited samples is an open problem in computer vision and biomedical imaging. Deep learning based vision systems mostly deal with high number of low-resolution images, whereas subtle detail in biomedical images require higher resolution. To bridge this gap, we propose a simple yet effective deep network that performs two tasks simultaneously in an end-to-end manner. First, it utilises a gated attention module that can focus on multiple key instances at high resolution without extra annotations or region proposals. Second, the global structural features and local instance features are fused for final image level classification. The result is a robust but lightweight end-to-end trainable deep network that yields state-of-the-art results in two separate fine-grained multi-instance biomedical image classification tasks: a benchmark breast cancer histology dataset and our new fungi species mycology dataset. In addition, we demonstrate the interpretability of the proposed model by visualising the concordance of the learned features with clinically relevant features.
Additive Angular Margin for Few Shot Learning to Classify Clinical Endoscopy Images
Mar 26, 2020
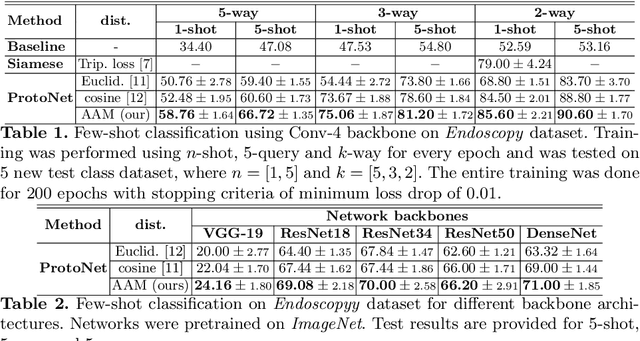
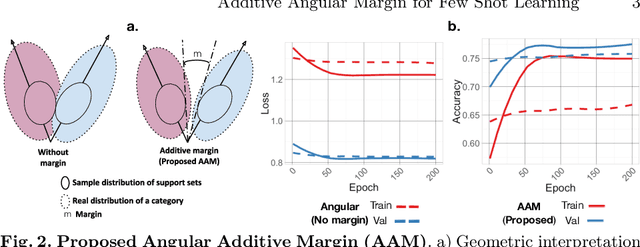
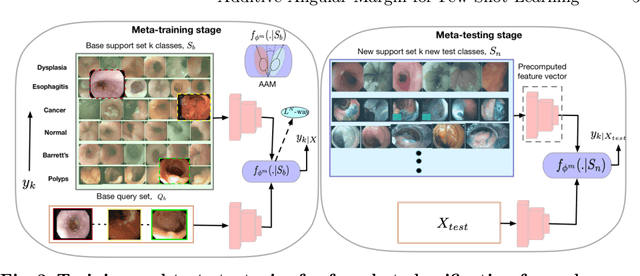
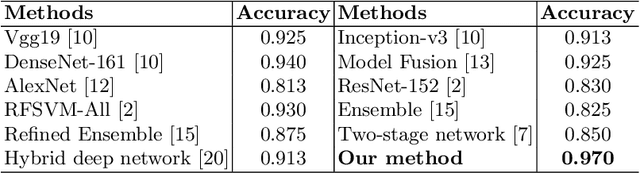
Abstract:Endoscopy is a widely used imaging modality to diagnose and treat diseases in hollow organs as for example the gastrointestinal tract, the kidney and the liver. However, due to varied modalities and use of different imaging protocols at various clinical centers impose significant challenges when generalising deep learning models. Moreover, the assembly of large datasets from different clinical centers can introduce a huge label bias that renders any learnt model unusable. Also, when using new modality or presence of images with rare patterns, a bulk amount of similar image data and their corresponding labels are required for training these models. In this work, we propose to use a few-shot learning approach that requires less training data and can be used to predict label classes of test samples from an unseen dataset. We propose a novel additive angular margin metric in the framework of prototypical network in few-shot learning setting. We compare our approach to the several established methods on a large cohort of multi-center, multi-organ, and multi-modal endoscopy data. The proposed algorithm outperforms existing state-of-the-art methods.
Endoscopy disease detection challenge 2020
Mar 07, 2020
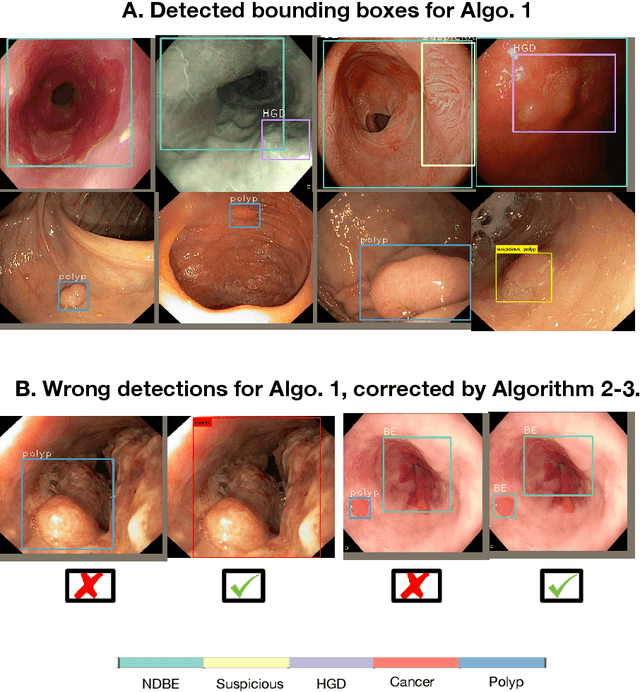
Abstract:Whilst many technologies are built around endoscopy, there is a need to have a comprehensive dataset collected from multiple centers to address the generalization issues with most deep learning frameworks. What could be more important than disease detection and localization? Through our extensive network of clinical and computational experts, we have collected, curated and annotated gastrointestinal endoscopy video frames. We have released this dataset and have launched disease detection and segmentation challenge EDD2020 https://edd2020.grand-challenge.org to address the limitations and explore new directions. EDD2020 is a crowd sourcing initiative to test the feasibility of recent deep learning methods and to promote research for building robust technologies. In this paper, we provide an overview of the EDD2020 dataset, challenge tasks, evaluation strategies and a short summary of results on test data. A detailed paper will be drafted after the challenge workshop with more detailed analysis of the results.
Semantic filtering through deep source separation on microscopy images
Sep 02, 2019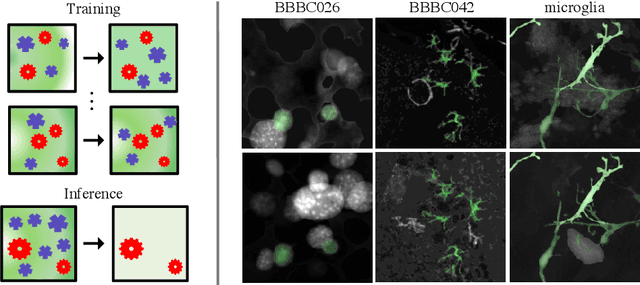
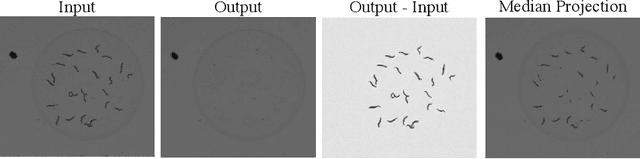
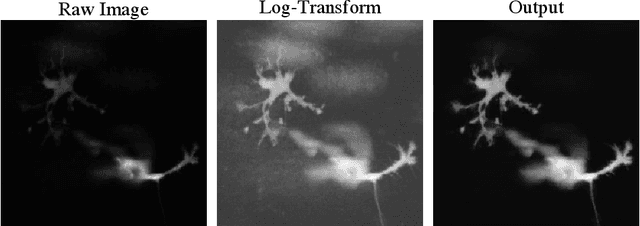
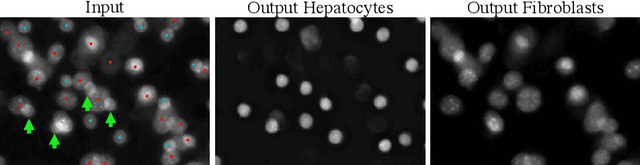
Abstract:By their very nature microscopy images of cells and tissues consist of a limited number of object types or components. In contrast to most natural scenes, the composition is known a priori. Decomposing biological images into semantically meaningful objects and layers is the aim of this paper. Building on recent approaches to image de-noising we present a framework that achieves state-of-the-art segmentation results requiring little or no manual annotations. Here, synthetic images generated by adding cell crops are sufficient to train the model. Extensive experiments on cellular images, a histology data set, and small animal videos demonstrate that our approach generalizes to a broad range of experimental settings. As the proposed methodology does not require densely labelled training images and is capable of resolving the partially overlapping objects it holds the promise of being of use in a number of different applications.
Conv2Warp: An unsupervised deformable image registration with continuous convolution and warping
Aug 16, 2019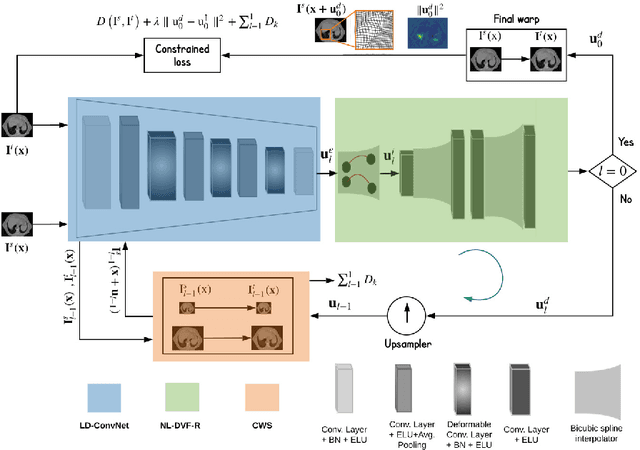
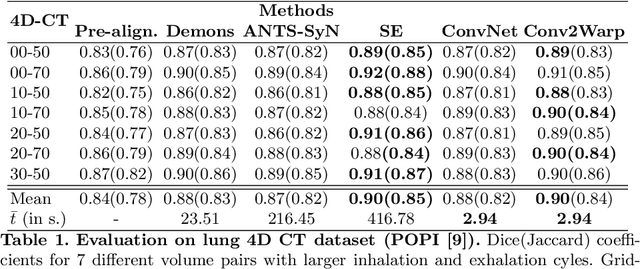
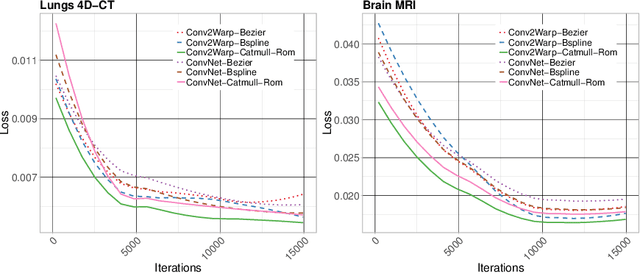
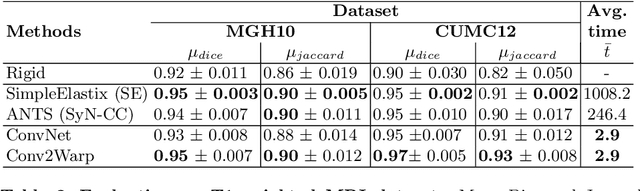
Abstract:Recent successes in deep learning based deformable image registration (DIR) methods have demonstrated that complex deformation can be learnt directly from data while reducing computation time when compared to traditional methods. However, the reliance on fully linear convolutional layers imposes a uniform sampling of pixel/voxel locations which ultimately limits their performance. To address this problem, we propose a novel approach of learning a continuous warp of the source image. Here, the required deformation vector fields are obtained from a concatenated linear and non-linear convolution layers and a learnable bicubic Catmull-Rom spline resampler. This allows to compute smooth deformation field and more accurate alignment compared to using only linear convolutions and linear resampling. In addition, the continuous warping technique penalizes disagreements that are due to topological changes. Our experiments demonstrate that this approach manages to capture large non-linear deformations and minimizes the propagation of interpolation errors. While improving accuracy the method is computationally efficient. We present comparative results on a range of public 4D CT lung (POPI) and brain datasets (CUMC12, MGH10).
Ink removal from histopathology whole slide images by combining classification, detection and image generation models
May 10, 2019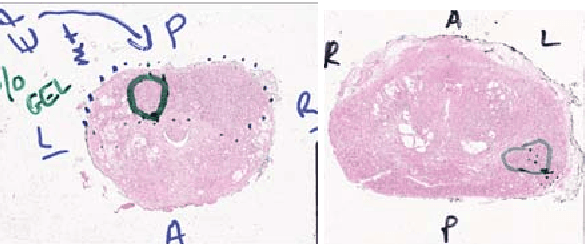
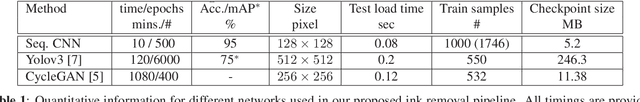

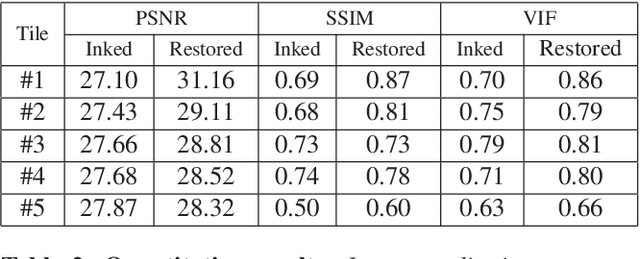
Abstract:Histopathology slides are routinely marked by pathologists using permanent ink markers that should not be removed as they form part of the medical record. Often tumour regions are marked up for the purpose of highlighting features or other downstream processing such an gene sequencing. Once digitised there is no established method for removing this information from the whole slide images limiting its usability in research and study. Removal of marker ink from these high-resolution whole slide images is non-trivial and complex problem as they contaminate different regions and in an inconsistent manner. We propose an efficient pipeline using convolution neural networks that results in ink-free images without compromising information and image resolution. Our pipeline includes a sequential classical convolution neural network for accurate classification of contaminated image tiles, a fast region detector and a domain adaptive cycle consistent adversarial generative model for restoration of foreground pixels. Both quantitative and qualitative results on four different whole slide images show that our approach yields visually coherent ink-free whole slide images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge