James H. F. Rudd
on behalf of the AIX-COVNET collaboration
SurvSurf: a partially monotonic neural network for first-hitting time prediction of intermittently observed discrete and continuous sequential events
Apr 07, 2025Abstract:We propose a neural-network based survival model (SurvSurf) specifically designed for direct and simultaneous probabilistic prediction of the first hitting time of sequential events from baseline. Unlike existing models, SurvSurf is theoretically guaranteed to never violate the monotonic relationship between the cumulative incidence functions of sequential events, while allowing nonlinear influence from predictors. It also incorporates implicit truths for unobserved intermediate events in model fitting, and supports both discrete and continuous time and events. We also identified a variant of the Integrated Brier Score (IBS) that showed robust correlation with the mean squared error (MSE) between the true and predicted probabilities by accounting for implied truths about the missing intermediate events. We demonstrated the superiority of SurvSurf compared to modern and traditional predictive survival models in two simulated datasets and two real-world datasets, using MSE, the more robust IBS and by measuring the extent of monotonicity violation.
Parameter choices in HaarPSI for IQA with medical images
Oct 31, 2024Abstract:When developing machine learning models, image quality assessment (IQA) measures are a crucial component for evaluation. However, commonly used IQA measures have been primarily developed and optimized for natural images. In many specialized settings, such as medical images, this poses an often-overlooked problem regarding suitability. In previous studies, the IQA measure HaarPSI showed promising behavior for natural and medical images. HaarPSI is based on Haar wavelet representations and the framework allows optimization of two parameters. So far, these parameters have been aligned for natural images. Here, we optimize these parameters for two annotated medical data sets, a photoacoustic and a chest X-Ray data set. We observe that they are more sensitive to the parameter choices than the employed natural images, and on the other hand both medical data sets lead to similar parameter values when optimized. We denote the optimized setting, which improves the performance for the medical images notably, by HaarPSI$_{MED}$. The results suggest that adapting common IQA measures within their frameworks for medical images can provide a valuable, generalizable addition to the employment of more specific task-based measures.
The curious case of the test set AUROC
Dec 19, 2023



Abstract:Whilst the size and complexity of ML models have rapidly and significantly increased over the past decade, the methods for assessing their performance have not kept pace. In particular, among the many potential performance metrics, the ML community stubbornly continues to use (a) the area under the receiver operating characteristic curve (AUROC) for a validation and test cohort (distinct from training data) or (b) the sensitivity and specificity for the test data at an optimal threshold determined from the validation ROC. However, we argue that considering scores derived from the test ROC curve alone gives only a narrow insight into how a model performs and its ability to generalise.
Recent Methodological Advances in Federated Learning for Healthcare
Oct 04, 2023



Abstract:For healthcare datasets, it is often not possible to combine data samples from multiple sites due to ethical, privacy or logistical concerns. Federated learning allows for the utilisation of powerful machine learning algorithms without requiring the pooling of data. Healthcare data has many simultaneous challenges which require new methodologies to address, such as highly-siloed data, class imbalance, missing data, distribution shifts and non-standardised variables. Federated learning adds significant methodological complexity to conventional centralised machine learning, requiring distributed optimisation, communication between nodes, aggregation of models and redistribution of models. In this systematic review, we consider all papers on Scopus that were published between January 2015 and February 2023 and which describe new federated learning methodologies for addressing challenges with healthcare data. We performed a detailed review of the 89 papers which fulfilled these criteria. Significant systemic issues were identified throughout the literature which compromise the methodologies in many of the papers reviewed. We give detailed recommendations to help improve the quality of the methodology development for federated learning in healthcare.
Reinterpreting survival analysis in the universal approximator age
Jul 25, 2023



Abstract:Survival analysis is an integral part of the statistical toolbox. However, while most domains of classical statistics have embraced deep learning, survival analysis only recently gained some minor attention from the deep learning community. This recent development is likely in part motivated by the COVID-19 pandemic. We aim to provide the tools needed to fully harness the potential of survival analysis in deep learning. On the one hand, we discuss how survival analysis connects to classification and regression. On the other hand, we provide technical tools. We provide a new loss function, evaluation metrics, and the first universal approximating network that provably produces survival curves without numeric integration. We show that the loss function and model outperform other approaches using a large numerical study.
Dis-AE: Multi-domain & Multi-task Generalisation on Real-World Clinical Data
Jun 15, 2023



Abstract:Clinical data is often affected by clinically irrelevant factors such as discrepancies between measurement devices or differing processing methods between sites. In the field of machine learning (ML), these factors are known as domains and the distribution differences they cause in the data are known as domain shifts. ML models trained using data from one domain often perform poorly when applied to data from another domain, potentially leading to wrong predictions. As such, developing machine learning models that can generalise well across multiple domains is a challenging yet essential task in the successful application of ML in clinical practice. In this paper, we propose a novel disentangled autoencoder (Dis-AE) neural network architecture that can learn domain-invariant data representations for multi-label classification of medical measurements even when the data is influenced by multiple interacting domain shifts at once. The model utilises adversarial training to produce data representations from which the domain can no longer be determined. We evaluate the model's domain generalisation capabilities on synthetic datasets and full blood count (FBC) data from blood donors as well as primary and secondary care patients, showing that Dis-AE improves model generalisation on multiple domains simultaneously while preserving clinically relevant information.
Classification of datasets with imputed missing values: does imputation quality matter?
Jun 16, 2022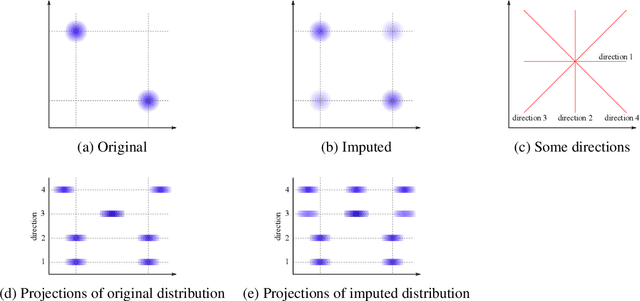

Abstract:Classifying samples in incomplete datasets is a common aim for machine learning practitioners, but is non-trivial. Missing data is found in most real-world datasets and these missing values are typically imputed using established methods, followed by classification of the now complete, imputed, samples. The focus of the machine learning researcher is then to optimise the downstream classification performance. In this study, we highlight that it is imperative to consider the quality of the imputation. We demonstrate how the commonly used measures for assessing quality are flawed and propose a new class of discrepancy scores which focus on how well the method recreates the overall distribution of the data. To conclude, we highlight the compromised interpretability of classifier models trained using poorly imputed data.
Machine learning for COVID-19 detection and prognostication using chest radiographs and CT scans: a systematic methodological review
Sep 01, 2020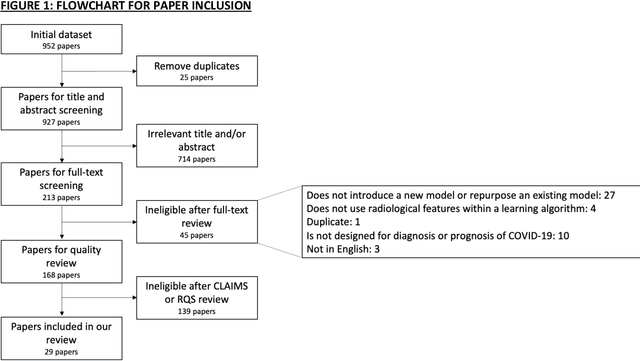
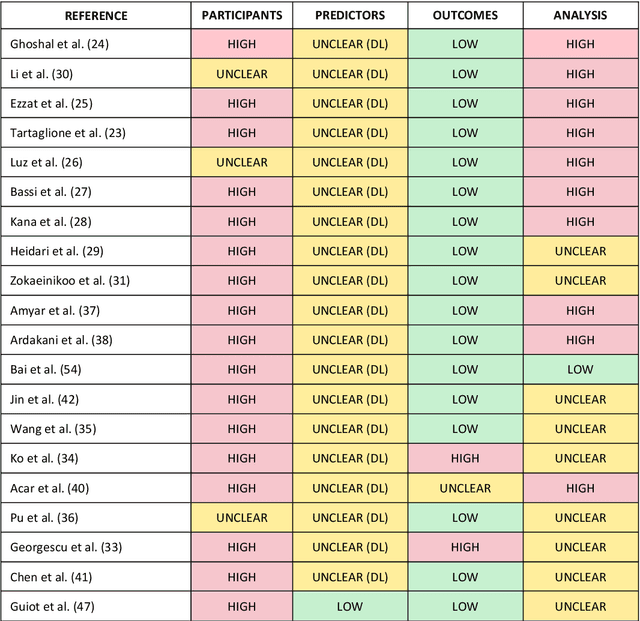
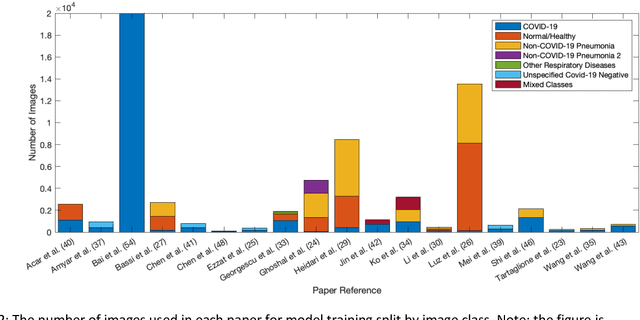
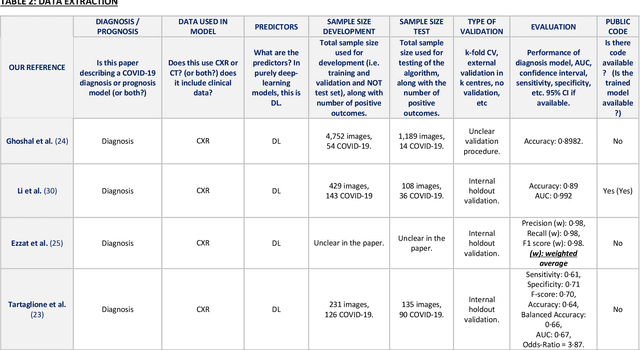
Abstract:Background: Machine learning methods offer great potential for fast and accurate detection and prognostication of COVID-19 from standard-of-care chest radiographs (CXR) and computed tomography (CT) images. In this systematic review we critically evaluate the machine learning methodologies employed in the rapidly growing literature. Methods: In this systematic review we reviewed EMBASE via OVID, MEDLINE via PubMed, bioRxiv, medRxiv and arXiv for published papers and preprints uploaded from Jan 1, 2020 to June 24, 2020. Studies which consider machine learning models for the diagnosis or prognosis of COVID-19 from CXR or CT images were included. A methodology quality review of each paper was performed against established benchmarks to ensure the review focusses only on high-quality reproducible papers. This study is registered with PROSPERO [CRD42020188887]. Interpretation: Our review finds that none of the developed models discussed are of potential clinical use due to methodological flaws and underlying biases. This is a major weakness, given the urgency with which validated COVID-19 models are needed. Typically, we find that the documentation of a model's development is not sufficient to make the results reproducible and therefore of 168 candidate papers only 29 are deemed to be reproducible and subsequently considered in this review. We therefore encourage authors to use established machine learning checklists to ensure sufficient documentation is made available, and to follow the PROBAST (prediction model risk of bias assessment tool) framework to determine the underlying biases in their model development process and to mitigate these where possible. This is key to safe clinical implementation which is urgently needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge