Cathal McCague
on behalf of the AIX-COVNET collaboration
A Self-Supervised Image Registration Approach for Measuring Local Response Patterns in Metastatic Ovarian Cancer
Jul 24, 2024



Abstract:High-grade serous ovarian carcinoma (HGSOC) is characterised by significant spatial and temporal heterogeneity, typically manifesting at an advanced metastatic stage. A major challenge in treating advanced HGSOC is effectively monitoring localised change in tumour burden across multiple sites during neoadjuvant chemotherapy (NACT) and predicting long-term pathological response and overall patient survival. In this work, we propose a self-supervised deformable image registration algorithm that utilises a general-purpose image encoder for image feature extraction to co-register contrast-enhanced computerised tomography scan images acquired before and after neoadjuvant chemotherapy. This approach addresses challenges posed by highly complex tumour deformations and longitudinal lesion matching during treatment. Localised tumour changes are calculated using the Jacobian determinant maps of the registration deformation at multiple disease sites and their macroscopic areas, including hypo-dense (i.e., cystic/necrotic), hyper-dense (i.e., calcified), and intermediate density (i.e., soft tissue) portions. A series of experiments is conducted to understand the role of a general-purpose image encoder and its application in quantifying change in tumour burden during neoadjuvant chemotherapy in HGSOC. This work is the first to demonstrate the feasibility of a self-supervised image registration approach in quantifying NACT-induced localised tumour changes across the whole disease burden of patients with complex multi-site HGSOC, which could be used as a potential marker for ovarian cancer patient's long-term pathological response and survival.
Calibrating Ensembles for Scalable Uncertainty Quantification in Deep Learning-based Medical Segmentation
Sep 20, 2022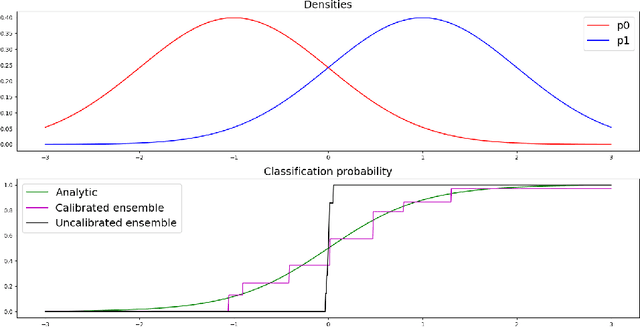
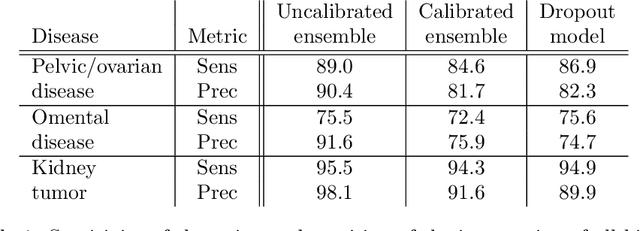
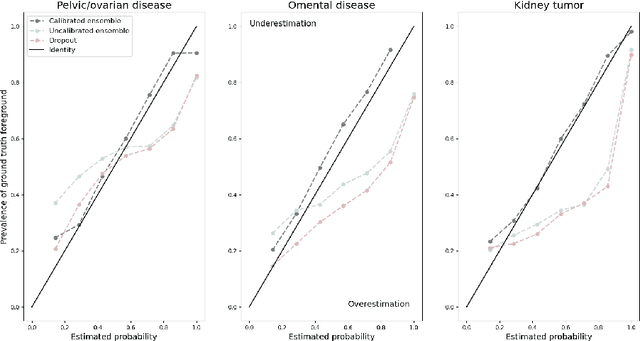
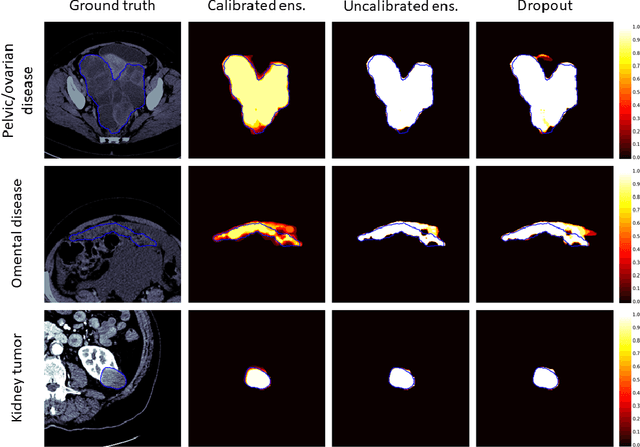
Abstract:Uncertainty quantification in automated image analysis is highly desired in many applications. Typically, machine learning models in classification or segmentation are only developed to provide binary answers; however, quantifying the uncertainty of the models can play a critical role for example in active learning or machine human interaction. Uncertainty quantification is especially difficult when using deep learning-based models, which are the state-of-the-art in many imaging applications. The current uncertainty quantification approaches do not scale well in high-dimensional real-world problems. Scalable solutions often rely on classical techniques, such as dropout, during inference or training ensembles of identical models with different random seeds to obtain a posterior distribution. In this paper, we show that these approaches fail to approximate the classification probability. On the contrary, we propose a scalable and intuitive framework to calibrate ensembles of deep learning models to produce uncertainty quantification measurements that approximate the classification probability. On unseen test data, we demonstrate improved calibration, sensitivity (in two out of three cases) and precision when being compared with the standard approaches. We further motivate the usage of our method in active learning, creating pseudo-labels to learn from unlabeled images and human-machine collaboration.
Machine learning for COVID-19 detection and prognostication using chest radiographs and CT scans: a systematic methodological review
Sep 01, 2020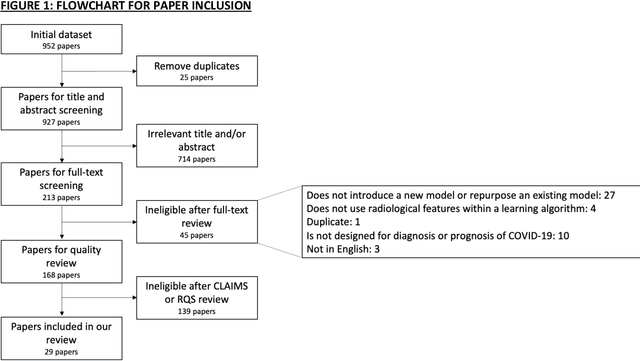
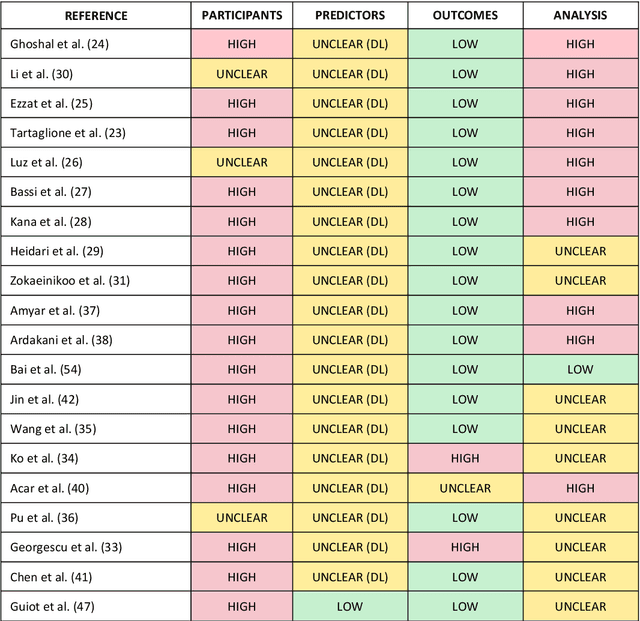
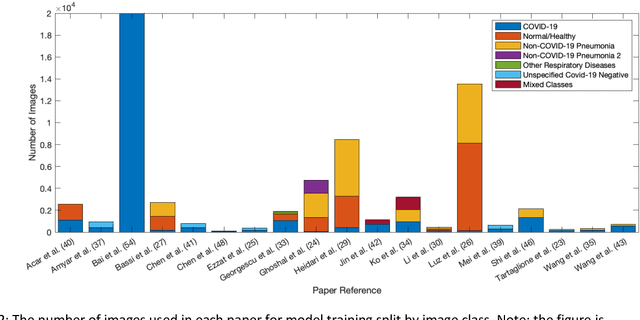
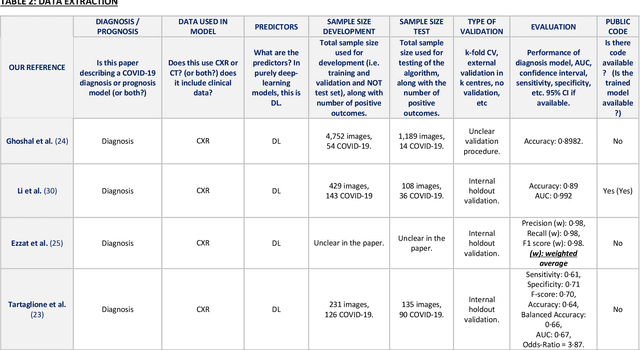
Abstract:Background: Machine learning methods offer great potential for fast and accurate detection and prognostication of COVID-19 from standard-of-care chest radiographs (CXR) and computed tomography (CT) images. In this systematic review we critically evaluate the machine learning methodologies employed in the rapidly growing literature. Methods: In this systematic review we reviewed EMBASE via OVID, MEDLINE via PubMed, bioRxiv, medRxiv and arXiv for published papers and preprints uploaded from Jan 1, 2020 to June 24, 2020. Studies which consider machine learning models for the diagnosis or prognosis of COVID-19 from CXR or CT images were included. A methodology quality review of each paper was performed against established benchmarks to ensure the review focusses only on high-quality reproducible papers. This study is registered with PROSPERO [CRD42020188887]. Interpretation: Our review finds that none of the developed models discussed are of potential clinical use due to methodological flaws and underlying biases. This is a major weakness, given the urgency with which validated COVID-19 models are needed. Typically, we find that the documentation of a model's development is not sufficient to make the results reproducible and therefore of 168 candidate papers only 29 are deemed to be reproducible and subsequently considered in this review. We therefore encourage authors to use established machine learning checklists to ensure sufficient documentation is made available, and to follow the PROBAST (prediction model risk of bias assessment tool) framework to determine the underlying biases in their model development process and to mitigate these where possible. This is key to safe clinical implementation which is urgently needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge