Jackie Ma
Metric Hub: A metric library and practical selection workflow for use-case-driven data quality assessment in medical AI
Jan 30, 2026Abstract:Machine learning (ML) in medicine has transitioned from research to concrete applications aimed at supporting several medical purposes like therapy selection, monitoring and treatment. Acceptance and effective adoption by clinicians and patients, as well as regulatory approval, require evidence of trustworthiness. A major factor for the development of trustworthy AI is the quantification of data quality for AI model training and testing. We have recently proposed the METRIC-framework for systematically evaluating the suitability (fit-for-purpose) of data for medical ML for a given task. Here, we operationalize this theoretical framework by introducing a collection of data quality metrics - the metric library - for practically measuring data quality dimensions. For each metric, we provide a metric card with the most important information, including definition, applicability, examples, pitfalls and recommendations, to support the understanding and implementation of these metrics. Furthermore, we discuss strategies and provide decision trees for choosing an appropriate set of data quality metrics from the metric library given specific use cases. We demonstrate the impact of our approach exemplarily on the PTB-XL ECG-dataset. This is a first step to enable fit-for-purpose evaluation of training and test data in practice as the base for establishing trustworthy AI in medicine.
Multimodal Deep Learning for Prediction of Progression-Free Survival in Patients with Neuroendocrine Tumors Undergoing 177Lu-based Peptide Receptor Radionuclide Therapy
Nov 07, 2025



Abstract:Peptide receptor radionuclide therapy (PRRT) is an established treatment for metastatic neuroendocrine tumors (NETs), yet long-term disease control occurs only in a subset of patients. Predicting progression-free survival (PFS) could support individualized treatment planning. This study evaluates laboratory, imaging, and multimodal deep learning models for PFS prediction in PRRT-treated patients. In this retrospective, single-center study 116 patients with metastatic NETs undergoing 177Lu-DOTATOC were included. Clinical characteristics, laboratory values, and pretherapeutic somatostatin receptor positron emission tomography/computed tomographies (SR-PET/CT) were collected. Seven models were trained to classify low- vs. high-PFS groups, including unimodal (laboratory, SR-PET, or CT) and multimodal fusion approaches. Explainability was evaluated by feature importance analysis and gradient maps. Forty-two patients (36%) had short PFS (< 1 year), 74 patients long PFS (>1 year). Groups were similar in most characteristics, except for higher baseline chromogranin A (p = 0.003), elevated gamma-GT (p = 0.002), and fewer PRRT cycles (p < 0.001) in short-PFS patients. The Random Forest model trained only on laboratory biomarkers reached an AUROC of 0.59 +- 0.02. Unimodal three-dimensional convolutional neural networks using SR-PET or CT performed worse (AUROC 0.42 +- 0.03 and 0.54 +- 0.01, respectively). A multimodal fusion model laboratory values, SR-PET, and CT -augmented with a pretrained CT branch - achieved the best results (AUROC 0.72 +- 0.01, AUPRC 0.80 +- 0.01). Multimodal deep learning combining SR-PET, CT, and laboratory biomarkers outperformed unimodal approaches for PFS prediction after PRRT. Upon external validation, such models may support risk-adapted follow-up strategies.
Spatially Resolved Meteorological and Ancillary Data in Central Europe for Rainfall Streamflow Modeling
Jun 04, 2025Abstract:We present a dataset for rainfall streamflow modeling that is fully spatially resolved with the aim of taking neural network-driven hydrological modeling beyond lumped catchments. To this end, we compiled data covering five river basins in central Europe: upper Danube, Elbe, Oder, Rhine, and Weser. The dataset contains meteorological forcings, as well as ancillary information on soil, rock, land cover, and orography. The data is harmonized to a regular 9km times 9km grid and contains daily values that span from October 1981 to September 2011. We also provide code to further combine our dataset with publicly available river discharge data for end-to-end rainfall streamflow modeling.
DiffScale: Continuous Downscaling and Bias Correction of Subseasonal Wind Speed Forecasts using Diffusion Models
Mar 31, 2025Abstract:Renewable resources are strongly dependent on local and large-scale weather situations. Skillful subseasonal to seasonal (S2S) forecasts -- beyond two weeks and up to two months -- can offer significant socioeconomic advantages to the energy sector. This study aims to enhance wind speed predictions using a diffusion model with classifier-free guidance to downscale S2S forecasts of surface wind speed. We propose DiffScale, a diffusion model that super-resolves spatial information for continuous downscaling factors and lead times. Leveraging weather priors as guidance for the generative process of diffusion models, we adopt the perspective of conditional probabilities on sampling super-resolved S2S forecasts. We aim to directly estimate the density associated with the target S2S forecasts at different spatial resolutions and lead times without auto-regression or sequence prediction, resulting in an efficient and flexible model. Synthetic experiments were designed to super-resolve wind speed S2S forecasts from the European Center for Medium-Range Weather Forecast (ECMWF) from a coarse resolution to a finer resolution of ERA5 reanalysis data, which serves as a high-resolution target. The innovative aspect of DiffScale lies in its flexibility to downscale arbitrary scaling factors, enabling it to generalize across various grid resolutions and lead times -without retraining the model- while correcting model errors, making it a versatile tool for improving S2S wind speed forecasts. We achieve a significant improvement in prediction quality, outperforming baselines up to week 3.
Synthetic Datasets for Machine Learning on Spatio-Temporal Graphs using PDEs
Feb 06, 2025Abstract:Many physical processes can be expressed through partial differential equations (PDEs). Real-world measurements of such processes are often collected at irregularly distributed points in space, which can be effectively represented as graphs; however, there are currently only a few existing datasets. Our work aims to make advancements in the field of PDE-modeling accessible to the temporal graph machine learning community, while addressing the data scarcity problem, by creating and utilizing datasets based on PDEs. In this work, we create and use synthetic datasets based on PDEs to support spatio-temporal graph modeling in machine learning for different applications. More precisely, we showcase three equations to model different types of disasters and hazards in the fields of epidemiology, atmospheric particles, and tsunami waves. Further, we show how such created datasets can be used by benchmarking several machine learning models on the epidemiological dataset. Additionally, we show how pre-training on this dataset can improve model performance on real-world epidemiological data. The presented methods enable others to create datasets and benchmarks customized to individual requirements. The source code for our methodology and the three created datasets can be found on https://github.com/github-usr-ano/Temporal_Graph_Data_PDEs.
Spatial Shortcuts in Graph Neural Controlled Differential Equations
Oct 25, 2024



Abstract:We incorporate prior graph topology information into a Neural Controlled Differential Equation (NCDE) to predict the future states of a dynamical system defined on a graph. The informed NCDE infers the future dynamics at the vertices of simulated advection data on graph edges with a known causal graph, observed only at vertices during training. We investigate different positions in the model architecture to inform the NCDE with graph information and identify an outer position between hidden state and control as theoretically and empirically favorable. Our such informed NCDE requires fewer parameters to reach a lower Mean Absolute Error (MAE) compared to previous methods that do not incorporate additional graph topology information.
Synthetic Generation of Dermatoscopic Images with GAN and Closed-Form Factorization
Oct 07, 2024



Abstract:In the realm of dermatological diagnoses, where the analysis of dermatoscopic and microscopic skin lesion images is pivotal for the accurate and early detection of various medical conditions, the costs associated with creating diverse and high-quality annotated datasets have hampered the accuracy and generalizability of machine learning models. We propose an innovative unsupervised augmentation solution that harnesses Generative Adversarial Network (GAN) based models and associated techniques over their latent space to generate controlled semiautomatically-discovered semantic variations in dermatoscopic images. We created synthetic images to incorporate the semantic variations and augmented the training data with these images. With this approach, we were able to increase the performance of machine learning models and set a new benchmark amongst non-ensemble based models in skin lesion classification on the HAM10000 dataset; and used the observed analytics and generated models for detailed studies on model explainability, affirming the effectiveness of our solution.
From CNNs to Vision Transformers -- A Comprehensive Evaluation of Deep Learning Models for Histopathology
Apr 11, 2022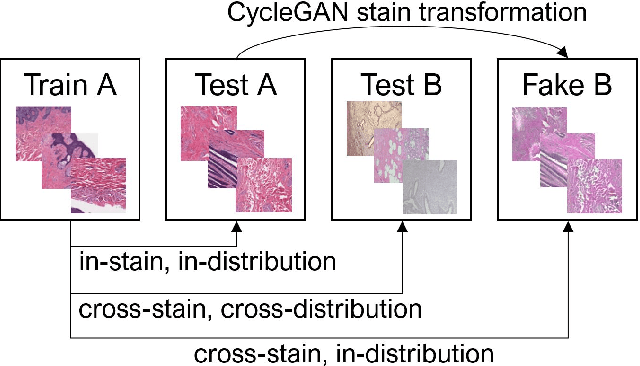
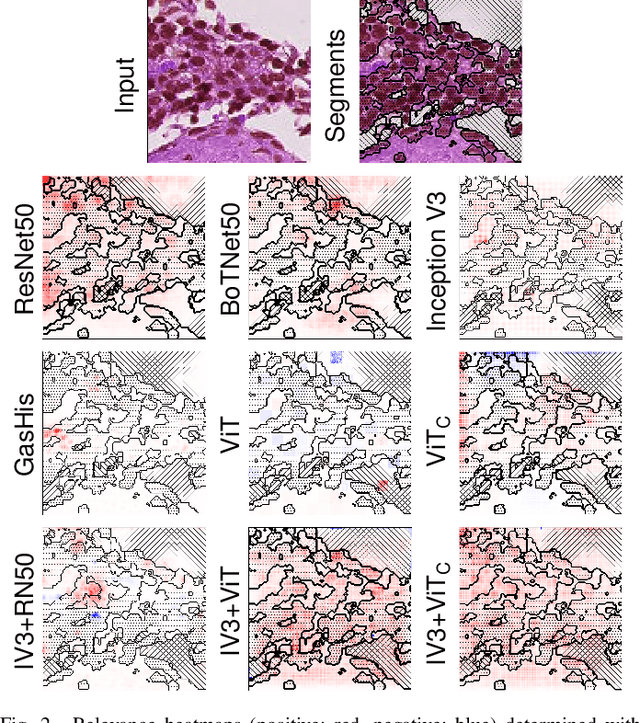
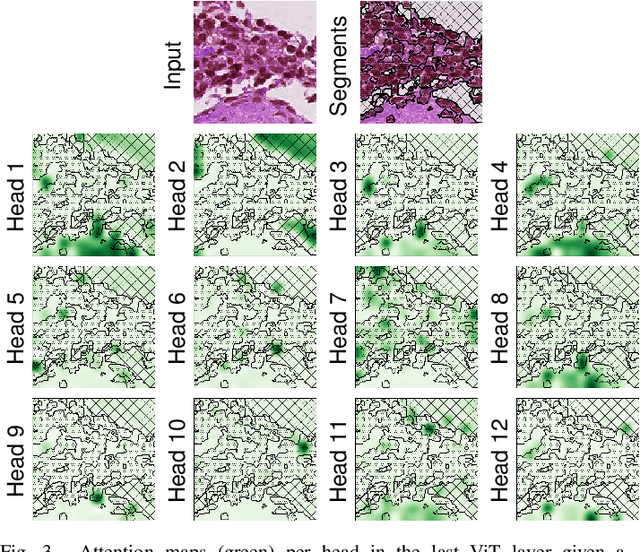
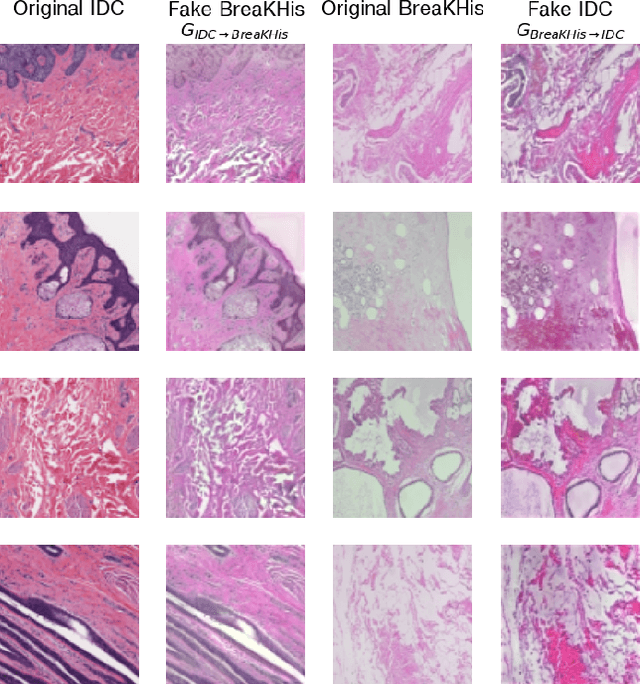
Abstract:While machine learning is currently transforming the field of histopathology, the domain lacks a comprehensive evaluation of state-of-the-art models based on essential but complementary quality requirements beyond a mere classification accuracy. In order to fill this gap, we conducted an extensive evaluation by benchmarking a wide range of classification models, including recent vision transformers, convolutional neural networks and hybrid models comprising transformer and convolutional models. We thoroughly tested the models on five widely used histopathology datasets containing whole slide images of breast, gastric, and colorectal cancer and developed a novel approach using an image-to-image translation model to assess the robustness of a cancer classification model against stain variations. Further, we extended existing interpretability methods to previously unstudied models and systematically reveal insights of the models' classification strategies that allow for plausibility checks and systematic comparisons. The study resulted in specific model recommendations for practitioners as well as putting forward a general methodology to quantify a model's quality according to complementary requirements that can be transferred to future model architectures.
On the Robustness of Pretraining and Self-Supervision for a Deep Learning-based Analysis of Diabetic Retinopathy
Jun 25, 2021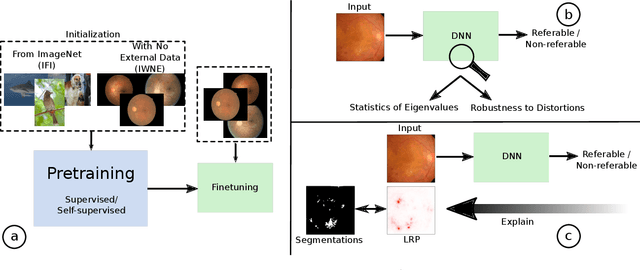

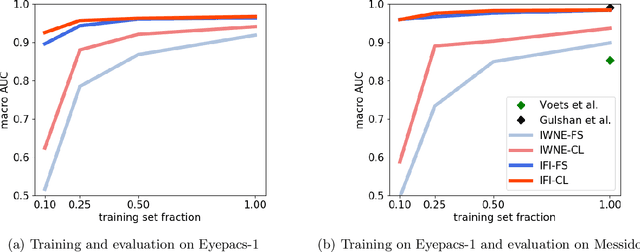
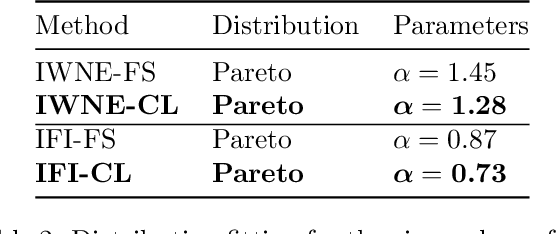
Abstract:There is an increasing number of medical use-cases where classification algorithms based on deep neural networks reach performance levels that are competitive with human medical experts. To alleviate the challenges of small dataset sizes, these systems often rely on pretraining. In this work, we aim to assess the broader implications of these approaches. For diabetic retinopathy grading as exemplary use case, we compare the impact of different training procedures including recently established self-supervised pretraining methods based on contrastive learning. To this end, we investigate different aspects such as quantitative performance, statistics of the learned feature representations, interpretability and robustness to image distortions. Our results indicate that models initialized from ImageNet pretraining report a significant increase in performance, generalization and robustness to image distortions. In particular, self-supervised models show further benefits to supervised models. Self-supervised models with initialization from ImageNet pretraining not only report higher performance, they also reduce overfitting to large lesions along with improvements in taking into account minute lesions indicative of the progression of the disease. Understanding the effects of pretraining in a broader sense that goes beyond simple performance comparisons is of crucial importance for the broader medical imaging community beyond the use-case considered in this work.
Risk Estimation of SARS-CoV-2 Transmission from Bluetooth Low Energy Measurements
Apr 22, 2020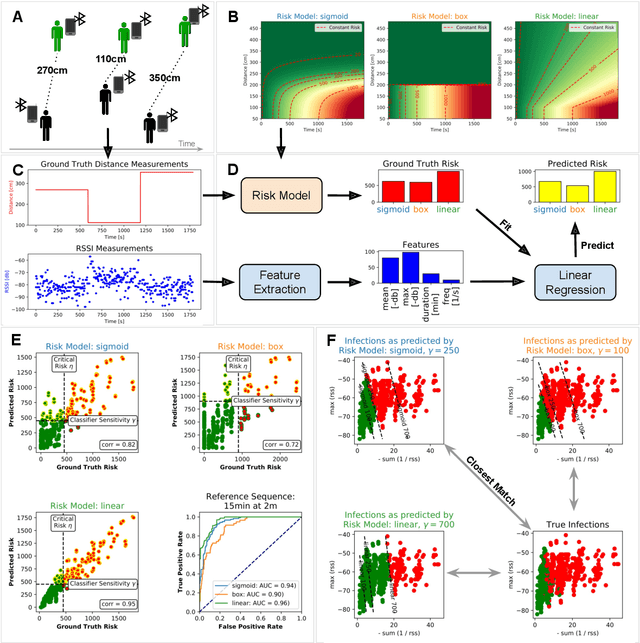
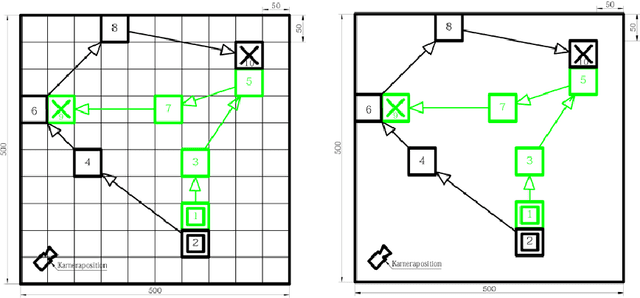
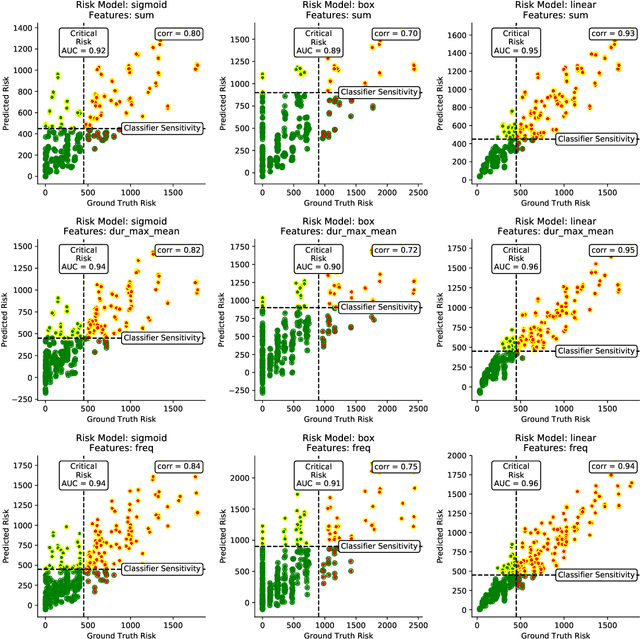
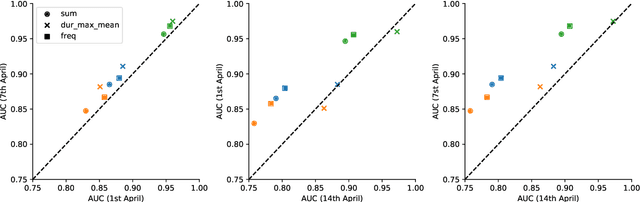
Abstract:Digital contact tracing approaches based on Bluetooth low energy (BLE) have the potential to efficiently contain and delay outbreaks of infectious diseases such as the ongoing SARS-CoV-2 pandemic. In this work we propose a novel machine learning based approach to reliably detect subjects that have spent enough time in close proximity to be at risk of being infected. Our study is an important proof of concept that will aid the battery of epidemiological policies aiming to slow down the rapid spread of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge