Klaus-Robert Müller
TUB
Investigating the Robustness of Subtask Distillation under Spurious Correlation
Jan 31, 2026Abstract:Subtask distillation is an emerging paradigm in which compact, specialized models are extracted from large, general-purpose 'foundation models' for deployment in environments with limited resources or in standalone computer systems. Although distillation uses a teacher model, it still relies on a dataset that is often limited in size and may lack representativeness or exhibit spurious correlations. In this paper, we evaluate established distillation methods, as well as the recent SubDistill method, when using data with spurious correlations for distillation. As the strength of the correlations increases, we observe a widening gap between advanced methods, such as SubDistill, which remain fairly robust, and some baseline methods, which degrade to near-random performance. Overall, our study underscores the challenges of knowledge distillation when applied to imperfect, real-world datasets, particularly those with spurious correlations.
Distilling Lightweight Domain Experts from Large ML Models by Identifying Relevant Subspaces
Jan 09, 2026Abstract:Knowledge distillation involves transferring the predictive capabilities of large, high-performing AI models (teachers) to smaller models (students) that can operate in environments with limited computing power. In this paper, we address the scenario in which only a few classes and their associated intermediate concepts are relevant to distill. This scenario is common in practice, yet few existing distillation methods explicitly focus on the relevant subtask. To address this gap, we introduce 'SubDistill', a new distillation algorithm with improved numerical properties that only distills the relevant components of the teacher model at each layer. Experiments on CIFAR-100 and ImageNet with Convolutional and Transformer models demonstrate that SubDistill outperforms existing layer-wise distillation techniques on a representative set of subtasks. Our benchmark evaluations are complemented by Explainable AI analyses showing that our distilled student models more closely match the decision structure of the original teacher model.
Atlas 2 - Foundation models for clinical deployment
Jan 08, 2026Abstract:Pathology foundation models substantially advanced the possibilities in computational pathology -- yet tradeoffs in terms of performance, robustness, and computational requirements remained, which limited their clinical deployment. In this report, we present Atlas 2, Atlas 2-B, and Atlas 2-S, three pathology vision foundation models which bridge these shortcomings by showing state-of-the-art performance in prediction performance, robustness, and resource efficiency in a comprehensive evaluation across eighty public benchmarks. Our models were trained on the largest pathology foundation model dataset to date comprising 5.5 million histopathology whole slide images, collected from three medical institutions Charité - Universtätsmedizin Berlin, LMU Munich, and Mayo Clinic.
Control Variate Score Matching for Diffusion Models
Dec 23, 2025Abstract:Diffusion models offer a robust framework for sampling from unnormalized probability densities, which requires accurately estimating the score of the noise-perturbed target distribution. While the standard Denoising Score Identity (DSI) relies on data samples, access to the target energy function enables an alternative formulation via the Target Score Identity (TSI). However, these estimators face a fundamental variance trade-off: DSI exhibits high variance in low-noise regimes, whereas TSI suffers from high variance at high noise levels. In this work, we reconcile these approaches by unifying both estimators within the principled framework of control variates. We introduce the Control Variate Score Identity (CVSI), deriving an optimal, time-dependent control coefficient that theoretically guarantees variance minimization across the entire noise spectrum. We demonstrate that CVSI serves as a robust, low-variance plug-in estimator that significantly enhances sample efficiency in both data-free sampler learning and inference-time diffusion sampling.
Noise & pattern: identity-anchored Tikhonov regularization for robust structural anomaly detection
Nov 10, 2025Abstract:Anomaly detection plays a pivotal role in automated industrial inspection, aiming to identify subtle or rare defects in otherwise uniform visual patterns. As collecting representative examples of all possible anomalies is infeasible, we tackle structural anomaly detection using a self-supervised autoencoder that learns to repair corrupted inputs. To this end, we introduce a corruption model that injects artificial disruptions into training images to mimic structural defects. While reminiscent of denoising autoencoders, our approach differs in two key aspects. First, instead of unstructured i.i.d.\ noise, we apply structured, spatially coherent perturbations that make the task a hybrid of segmentation and inpainting. Second, and counterintuitively, we add and preserve Gaussian noise on top of the occlusions, which acts as a Tikhonov regularizer anchoring the Jacobian of the reconstruction function toward identity. This identity-anchored regularization stabilizes reconstruction and further improves both detection and segmentation accuracy. On the MVTec AD benchmark, our method achieves state-of-the-art results (I/P-AUROC: 99.9/99.4), supporting our theoretical framework and demonstrating its practical relevance for automatic inspection.
Fast and Accurate Explanations of Distance-Based Classifiers by Uncovering Latent Explanatory Structures
Aug 05, 2025Abstract:Distance-based classifiers, such as k-nearest neighbors and support vector machines, continue to be a workhorse of machine learning, widely used in science and industry. In practice, to derive insights from these models, it is also important to ensure that their predictions are explainable. While the field of Explainable AI has supplied methods that are in principle applicable to any model, it has also emphasized the usefulness of latent structures (e.g. the sequence of layers in a neural network) to produce explanations. In this paper, we contribute by uncovering a hidden neural network structure in distance-based classifiers (consisting of linear detection units combined with nonlinear pooling layers) upon which Explainable AI techniques such as layer-wise relevance propagation (LRP) become applicable. Through quantitative evaluations, we demonstrate the advantage of our novel explanation approach over several baselines. We also show the overall usefulness of explaining distance-based models through two practical use cases.
Towards Robust Foundation Models for Digital Pathology
Jul 22, 2025
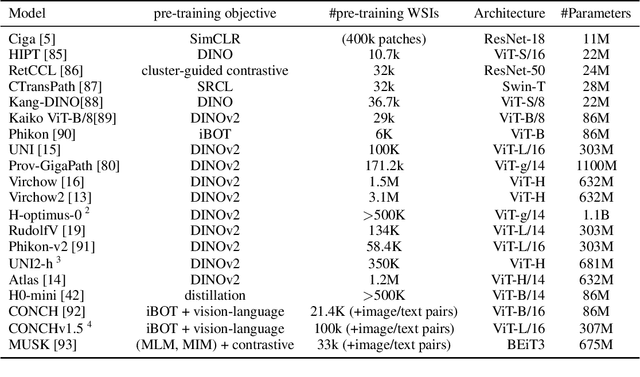
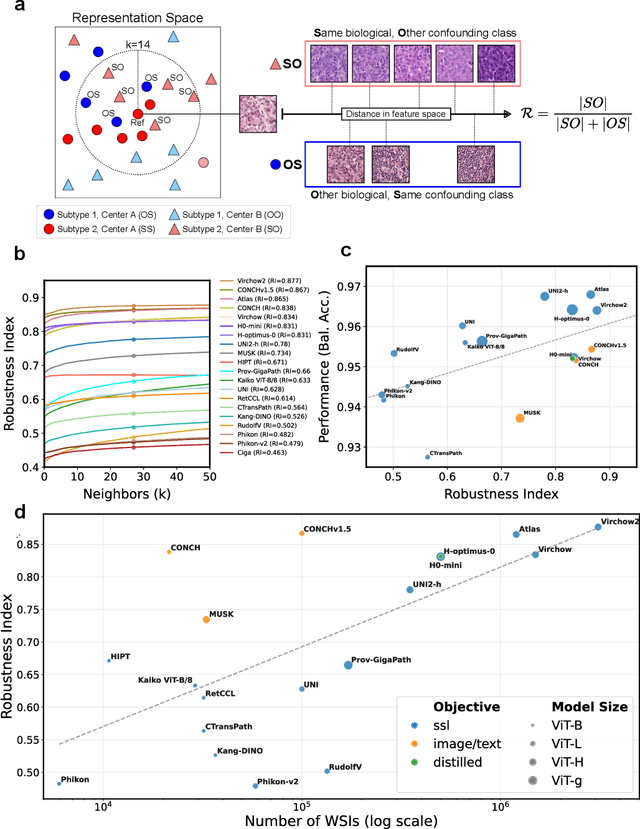

Abstract:Biomedical Foundation Models (FMs) are rapidly transforming AI-enabled healthcare research and entering clinical validation. However, their susceptibility to learning non-biological technical features -- including variations in surgical/endoscopic techniques, laboratory procedures, and scanner hardware -- poses risks for clinical deployment. We present the first systematic investigation of pathology FM robustness to non-biological features. Our work (i) introduces measures to quantify FM robustness, (ii) demonstrates the consequences of limited robustness, and (iii) proposes a framework for FM robustification to mitigate these issues. Specifically, we developed PathoROB, a robustness benchmark with three novel metrics, including the robustness index, and four datasets covering 28 biological classes from 34 medical centers. Our experiments reveal robustness deficits across all 20 evaluated FMs, and substantial robustness differences between them. We found that non-robust FM representations can cause major diagnostic downstream errors and clinical blunders that prevent safe clinical adoption. Using more robust FMs and post-hoc robustification considerably reduced (but did not yet eliminate) the risk of such errors. This work establishes that robustness evaluation is essential for validating pathology FMs before clinical adoption and demonstrates that future FM development must integrate robustness as a core design principle. PathoROB provides a blueprint for assessing robustness across biomedical domains, guiding FM improvement efforts towards more robust, representative, and clinically deployable AI systems that prioritize biological information over technical artifacts.
Sampling 3D Molecular Conformers with Diffusion Transformers
Jun 18, 2025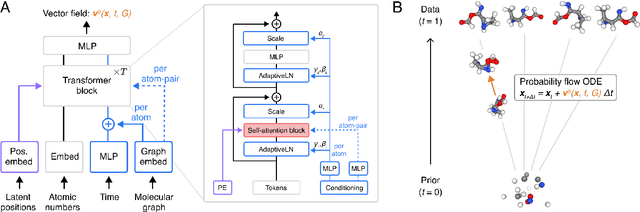
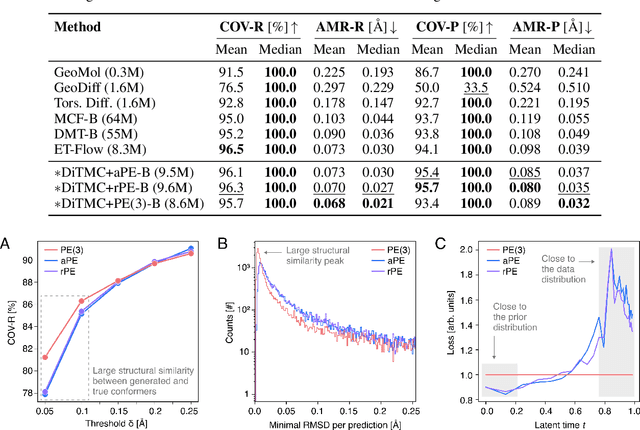
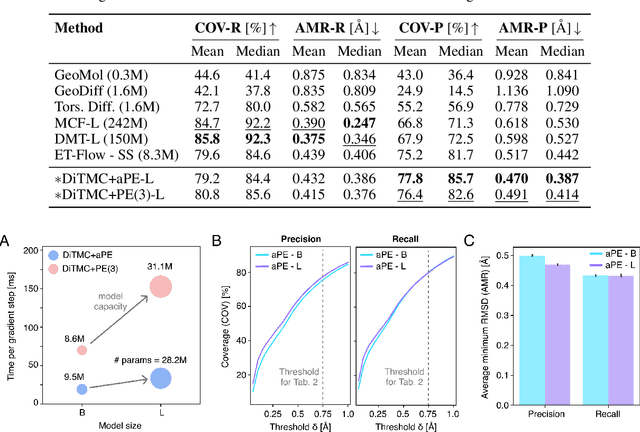
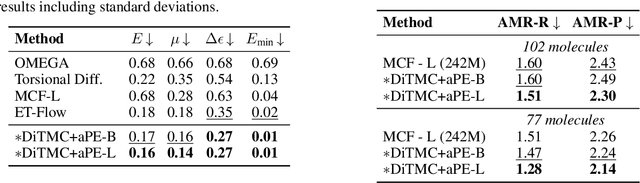
Abstract:Diffusion Transformers (DiTs) have demonstrated strong performance in generative modeling, particularly in image synthesis, making them a compelling choice for molecular conformer generation. However, applying DiTs to molecules introduces novel challenges, such as integrating discrete molecular graph information with continuous 3D geometry, handling Euclidean symmetries, and designing conditioning mechanisms that generalize across molecules of varying sizes and structures. We propose DiTMC, a framework that adapts DiTs to address these challenges through a modular architecture that separates the processing of 3D coordinates from conditioning on atomic connectivity. To this end, we introduce two complementary graph-based conditioning strategies that integrate seamlessly with the DiT architecture. These are combined with different attention mechanisms, including both standard non-equivariant and SO(3)-equivariant formulations, enabling flexible control over the trade-off between between accuracy and computational efficiency. Experiments on standard conformer generation benchmarks (GEOM-QM9, -DRUGS, -XL) demonstrate that DiTMC achieves state-of-the-art precision and physical validity. Our results highlight how architectural choices and symmetry priors affect sample quality and efficiency, suggesting promising directions for large-scale generative modeling of molecular structures. Code available at https://github.com/ML4MolSim/dit_mc.
Towards Desiderata-Driven Design of Visual Counterfactual Explainers
Jun 17, 2025Abstract:Visual counterfactual explainers (VCEs) are a straightforward and promising approach to enhancing the transparency of image classifiers. VCEs complement other types of explanations, such as feature attribution, by revealing the specific data transformations to which a machine learning model responds most strongly. In this paper, we argue that existing VCEs focus too narrowly on optimizing sample quality or change minimality; they fail to consider the more holistic desiderata for an explanation, such as fidelity, understandability, and sufficiency. To address this shortcoming, we explore new mechanisms for counterfactual generation and investigate how they can help fulfill these desiderata. We combine these mechanisms into a novel 'smooth counterfactual explorer' (SCE) algorithm and demonstrate its effectiveness through systematic evaluations on synthetic and real data.
Uncovering the Structure of Explanation Quality with Spectral Analysis
Apr 11, 2025Abstract:As machine learning models are increasingly considered for high-stakes domains, effective explanation methods are crucial to ensure that their prediction strategies are transparent to the user. Over the years, numerous metrics have been proposed to assess quality of explanations. However, their practical applicability remains unclear, in particular due to a limited understanding of which specific aspects each metric rewards. In this paper we propose a new framework based on spectral analysis of explanation outcomes to systematically capture the multifaceted properties of different explanation techniques. Our analysis uncovers two distinct factors of explanation quality-stability and target sensitivity-that can be directly observed through spectral decomposition. Experiments on both MNIST and ImageNet show that popular evaluation techniques (e.g., pixel-flipping, entropy) partially capture the trade-offs between these factors. Overall, our framework provides a foundational basis for understanding explanation quality, guiding the development of more reliable techniques for evaluating explanations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge