Harrisen Scells
Topic-Specific Classifiers are Better Relevance Judges than Prompted LLMs
Oct 06, 2025Abstract:The unjudged document problem, where pooled test collections have incomplete relevance judgments for evaluating new retrieval systems, is a key obstacle to the reusability of test collections in information retrieval. While the de facto standard to deal with the problem is to treat unjudged documents as non-relevant, many alternatives have been proposed, including the use of large language models (LLMs) as a relevance judge (LLM-as-a-judge). However, this has been criticized as circular, since the same LLM can be used as a judge and as a ranker at the same time. We propose to train topic-specific relevance classifiers instead: By finetuning monoT5 with independent LoRA weight adaptation on the judgments of a single assessor for a single topic's pool, we align it to that assessor's notion of relevance for the topic. The system rankings obtained through our classifier's relevance judgments achieve a Spearmans' $\rho$ correlation of $>0.95$ with ground truth system rankings. As little as 128 initial human judgments per topic suffice to improve the comparability of models, compared to treating unjudged documents as non-relevant, while achieving more reliability than existing LLM-as-a-judge approaches. Topic-specific relevance classifiers thus are a lightweight and straightforward way to tackle the unjudged document problem, while maintaining human judgments as the gold standard for retrieval evaluation. Code, models, and data are made openly available.
Reassessing Large Language Model Boolean Query Generation for Systematic Reviews
May 12, 2025



Abstract:Systematic reviews are comprehensive literature reviews that address highly focused research questions and represent the highest form of evidence in medicine. A critical step in this process is the development of complex Boolean queries to retrieve relevant literature. Given the difficulty of manually constructing these queries, recent efforts have explored Large Language Models (LLMs) to assist in their formulation. One of the first studies,Wang et al., investigated ChatGPT for this task, followed by Staudinger et al., which evaluated multiple LLMs in a reproducibility study. However, the latter overlooked several key aspects of the original work, including (i) validation of generated queries, (ii) output formatting constraints, and (iii) selection of examples for chain-of-thought (Guided) prompting. As a result, its findings diverged significantly from the original study. In this work, we systematically reproduce both studies while addressing these overlooked factors. Our results show that query effectiveness varies significantly across models and prompt designs, with guided query formulation benefiting from well-chosen seed studies. Overall, prompt design and model selection are key drivers of successful query formulation. Our findings provide a clearer understanding of LLMs' potential in Boolean query generation and highlight the importance of model- and prompt-specific optimisations. The complex nature of systematic reviews adds to challenges in both developing and reproducing methods but also highlights the importance of reproducibility studies in this domain.
The Viability of Crowdsourcing for RAG Evaluation
Apr 22, 2025



Abstract:How good are humans at writing and judging responses in retrieval-augmented generation (RAG) scenarios? To answer this question, we investigate the efficacy of crowdsourcing for RAG through two complementary studies: response writing and response utility judgment. We present the Crowd RAG Corpus 2025 (CrowdRAG-25), which consists of 903 human-written and 903 LLM-generated responses for the 301 topics of the TREC RAG'24 track, across the three discourse styles 'bulleted list', 'essay', and 'news'. For a selection of 65 topics, the corpus further contains 47,320 pairwise human judgments and 10,556 pairwise LLM judgments across seven utility dimensions (e.g., coverage and coherence). Our analyses give insights into human writing behavior for RAG and the viability of crowdsourcing for RAG evaluation. Human pairwise judgments provide reliable and cost-effective results compared to LLM-based pairwise or human/LLM-based pointwise judgments, as well as automated comparisons with human-written reference responses. All our data and tools are freely available.
AiReview: An Open Platform for Accelerating Systematic Reviews with LLMs
Apr 05, 2025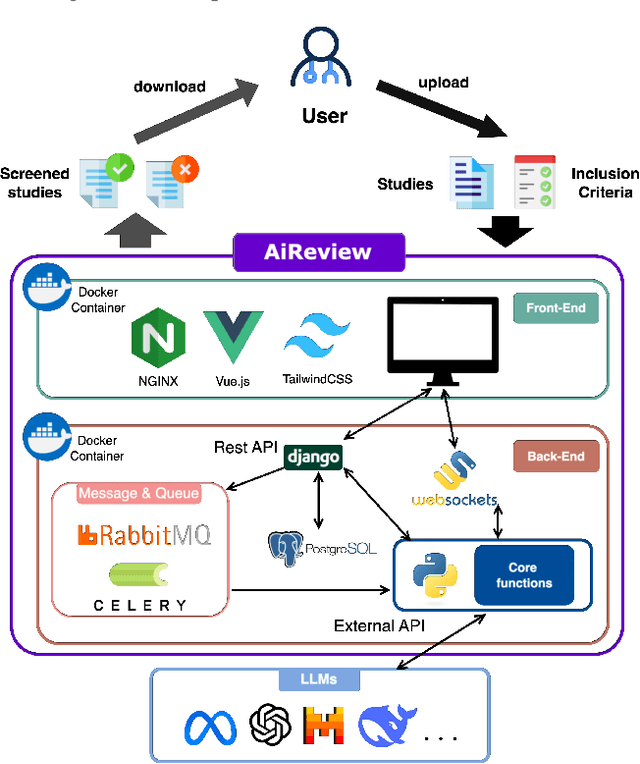
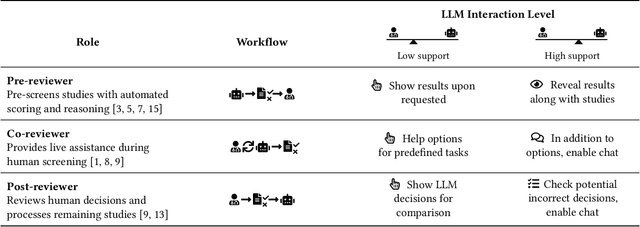
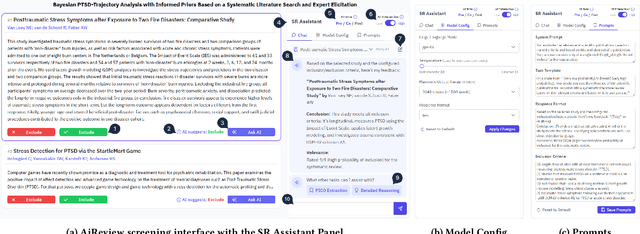
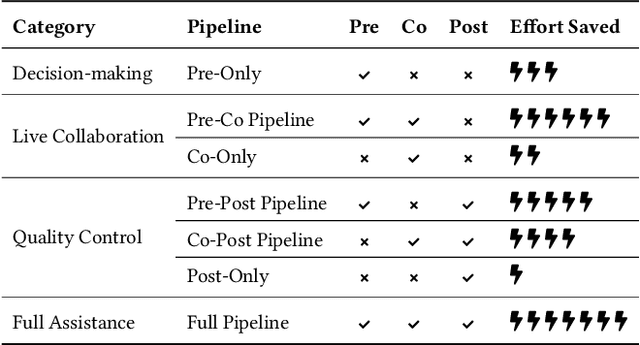
Abstract:Systematic reviews are fundamental to evidence-based medicine. Creating one is time-consuming and labour-intensive, mainly due to the need to screen, or assess, many studies for inclusion in the review. Several tools have been developed to streamline this process, mostly relying on traditional machine learning methods. Large language models (LLMs) have shown potential in further accelerating the screening process. However, no tool currently allows end users to directly leverage LLMs for screening or facilitates systematic and transparent usage of LLM-assisted screening methods. This paper introduces (i) an extensible framework for applying LLMs to systematic review tasks, particularly title and abstract screening, and (ii) a web-based interface for LLM-assisted screening. Together, these elements form AiReview-a novel platform for LLM-assisted systematic review creation. AiReview is the first of its kind to bridge the gap between cutting-edge LLM-assisted screening methods and those that create medical systematic reviews. The tool is available at https://aireview.ielab.io. The source code is also open sourced at https://github.com/ielab/ai-review.
Variations in Relevance Judgments and the Shelf Life of Test Collections
Feb 28, 2025Abstract:The fundamental property of Cranfield-style evaluations, that system rankings are stable even when assessors disagree on individual relevance decisions, was validated on traditional test collections. However, the paradigm shift towards neural retrieval models affected the characteristics of modern test collections, e.g., documents are short, judged with four grades of relevance, and information needs have no descriptions or narratives. Under these changes, it is unclear whether assessor disagreement remains negligible for system comparisons. We investigate this aspect under the additional condition that the few modern test collections are heavily re-used. Given more possible query interpretations due to less formalized information needs, an ''expiration date'' for test collections might be needed if top-effectiveness requires overfitting to a single interpretation of relevance. We run a reproducibility study and re-annotate the relevance judgments of the 2019 TREC Deep Learning track. We can reproduce prior work in the neural retrieval setting, showing that assessor disagreement does not affect system rankings. However, we observe that some models substantially degrade with our new relevance judgments, and some have already reached the effectiveness of humans as rankers, providing evidence that test collections can expire.
DenseReviewer: A Screening Prioritisation Tool for Systematic Review based on Dense Retrieval
Feb 05, 2025Abstract:Screening is a time-consuming and labour-intensive yet required task for medical systematic reviews, as tens of thousands of studies often need to be screened. Prioritising relevant studies to be screened allows downstream systematic review creation tasks to start earlier and save time. In previous work, we developed a dense retrieval method to prioritise relevant studies with reviewer feedback during the title and abstract screening stage. Our method outperforms previous active learning methods in both effectiveness and efficiency. In this demo, we extend this prior work by creating (1) a web-based screening tool that enables end-users to screen studies exploiting state-of-the-art methods and (2) a Python library that integrates models and feedback mechanisms and allows researchers to develop and demonstrate new active learning methods. We describe the tool's design and showcase how it can aid screening. The tool is available at https://densereviewer.ielab.io. The source code is also open sourced at https://github.com/ielab/densereviewer.
Ranking Generated Answers: On the Agreement of Retrieval Models with Humans on Consumer Health Questions
Aug 19, 2024

Abstract:Evaluating the output of generative large language models (LLMs) is challenging and difficult to scale. Most evaluations of LLMs focus on tasks such as single-choice question-answering or text classification. These tasks are not suitable for assessing open-ended question-answering capabilities, which are critical in domains where expertise is required, such as health, and where misleading or incorrect answers can have a significant impact on a user's health. Using human experts to evaluate the quality of LLM answers is generally considered the gold standard, but expert annotation is costly and slow. We present a method for evaluating LLM answers that uses ranking signals as a substitute for explicit relevance judgements. Our scoring method correlates with the preferences of human experts. We validate it by investigating the well-known fact that the quality of generated answers improves with the size of the model as well as with more sophisticated prompting strategies.
Learning Effective Representations for Retrieval Using Self-Distillation with Adaptive Relevance Margins
Jul 31, 2024



Abstract:Representation-based retrieval models, so-called biencoders, estimate the relevance of a document to a query by calculating the similarity of their respective embeddings. Current state-of-the-art biencoders are trained using an expensive training regime involving knowledge distillation from a teacher model and batch-sampling. Instead of relying on a teacher model, we contribute a novel parameter-free loss function for self-supervision that exploits the pre-trained language modeling capabilities of the encoder model as a training signal, eliminating the need for batch sampling by performing implicit hard negative mining. We investigate the capabilities of our proposed approach through extensive ablation studies, demonstrating that self-distillation can match the effectiveness of teacher distillation using only 13.5% of the data, while offering a speedup in training time between 3x and 15x compared to parametrized losses. Code and data is made openly available.
A Systematic Investigation of Distilling Large Language Models into Cross-Encoders for Passage Re-ranking
May 13, 2024


Abstract:Cross-encoders distilled from large language models are more effective re-rankers than cross-encoders fine-tuned using manually labeled data. However, the distilled models do not reach the language model's effectiveness. We construct and release a new distillation dataset, named Rank-DistiLLM, to investigate whether insights from fine-tuning cross-encoders on manually labeled data -- hard-negative sampling, deep sampling, and listwise loss functions -- are transferable to large language model ranker distillation. Our dataset can be used to train cross-encoders that reach the effectiveness of large language models while being orders of magnitude more efficient. Code and data is available at: https://github.com/webis-de/msmarco-llm-distillation
Set-Encoder: Permutation-Invariant Inter-Passage Attention for Listwise Passage Re-Ranking with Cross-Encoders
Apr 11, 2024Abstract:Cross-encoders are effective passage re-rankers. But when re-ranking multiple passages at once, existing cross-encoders inefficiently optimize the output ranking over several input permutations, as their passage interactions are not permutation-invariant. Moreover, their high memory footprint constrains the number of passages during listwise training. To tackle these issues, we propose the Set-Encoder, a new cross-encoder architecture that (1) introduces inter-passage attention with parallel passage processing to ensure permutation invariance between input passages, and that (2) uses fused-attention kernels to enable training with more passages at a time. In experiments on TREC Deep Learning and TIREx, the Set-Encoder is more effective than previous cross-encoders with a similar number of parameters. Compared to larger models, the Set-Encoder is more efficient and either on par or even more effective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge